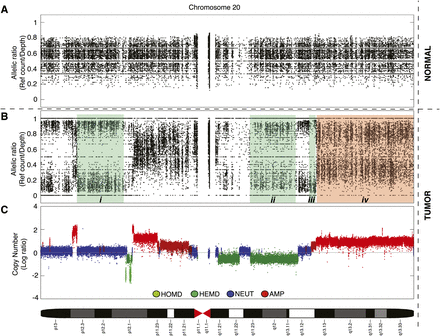
Illustration of empirical allelic ratios between tumor and normal genomic sequencing data from chromosome 20 of a triple-negative breast cancer genome (SA225), and effects of copy number. (A) Allelic ratio data of heterozygous loci in the normal genome are centered around 0.5, which represents the presence of two alleles. (B) At the same corresponding loci, allelic ratios in the tumor genome reveal four examples of somatically acquired segments of allelic imbalance in regions (i)–(iv). (C) The segmental copy number of the tumor helps give context to the allelic data: (i) copy neutral LOH (NLOH), AA/BB; (ii) deletion-induced LOH (DLOH), A/B; (iii) amplified LOH (ALOH), AAA/BBB; and (iv) allele-specific amplification (ASCNA), AAAB/ABBB. Allelic ratio value is defined as the reference read counts divided by total depth at a given position. A and B represent reference and nonreference alleles in the genotype, respectively.











