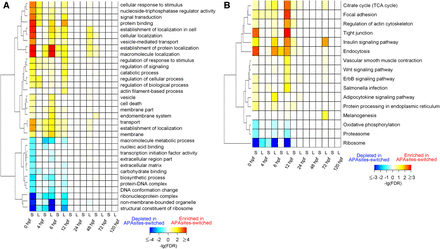
Functional annotations of APAsites-switching genes. (A) Gene ontology (GO) terms significantly (P < 0.011; 10% false discovery rate) enriched and depleted in APAsites-switching genes by taking single-UTR genes as the background across developmental stages. GO terms enriched in APAsites-switching genes are assigned a positive P-value (hence, positive FDR in the BH sense), and depleted terms are assigned a negative P-value (hence, negative FDR). (S) Genes with 3′ UTRs shortened; (L) genes with 3′ UTRs lengthened. (B) KEGG terms significantly (P < 0.013; 20% FDR) enriched and depleted in APAsites-switching gene. Data are presented as in A.











