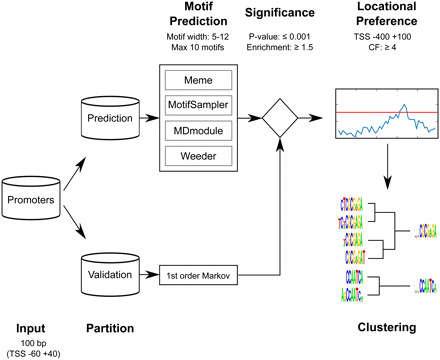
Schematic representation of the systematic de novo motif discovery pipeline. A set of input sequences is partitioned into two sets: a prediction set and a validation set. The prediction set is used as input for several different motif prediction algorithms. The validation set is used to produce a background set of random sequences generated with a first-order Markov model trained on the validation sequences. All predicted motifs are filtered for significance based on the hypergeometric distribution in the validation sequences compared with the random sequences. Only significant motifs with a positional bias, determined using the clustering factor, are kept. Subsequently this set of redundant motifs is clustered using an iterative procedure incorporating the new weighted information content (WIC) motif similarity score. To predict Xenopus promoter motifs, this pipeline was repeated 10 times.











