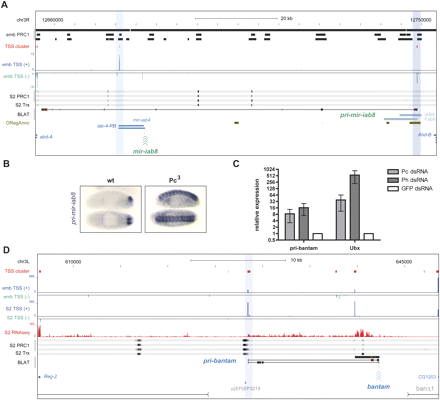
PcG proteins regulate transcription of pri-mir-iab8 and pri-bantam. (A) Screenshot from the UCSC Genome Browser covering the intergenic region between Abd-B and abd-A. TSS clusters at core promoter of pri-mir-iab8 (BLAT result, black) and iab4-RB locate to embryonic PcG binding sites (“emb PRC1”) (Schuettengruber et al. 2009). Fab-8 region (chr3R:12,740,929–12,748,923) (Barges et al. 2000), IAB8 enhancer (chr3R:12,744,584–12,749,967) (Zhou et al. 1999), and transcription factor binding sites (ORegAnno) (Griffith et al. 2008) as indicated. (B) In situ hybridization on whole-mount Drosophila embryos show a derepression of pri-mir-iab8 in homozygous Pc[3] mutant embryos. (C) S2 cells treated with dsRNAs against PRC1 components show derepression of the PcG target genes Ubx and pri-bantam. (D) Screenshot covering the intergenic region between Reg-2 and CG12030. The core promoter of the ∼20-kb pri-bantam transcript is bound by PRC1 and TRX proteins in S2 cells. Three primary transcripts found are shown as BLAT result and boundaries of known lesions and insertions are indicated (Hipfner et al. 2002).











