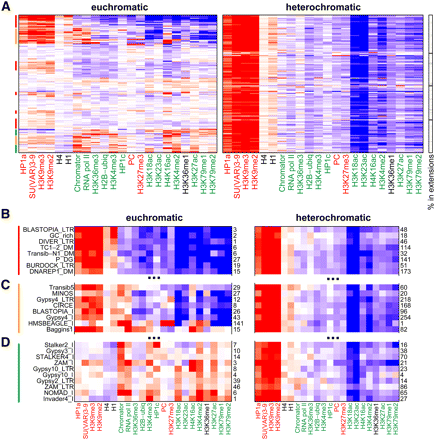
Repetitive elements integrated within heterochromatic regions show similar epigenomic signatures. (A) Average enrichments (red) and depletions (blue) for particular chromatin marks (columns) in BG3 cells are shown for specific repetitive element types (rows) in euchromatic (left) and heterochromatic (right) regions (extended version with repeat names is shown in Supplemental Fig. 11 for BG3 cells, and Supplemental Fig. 12 for S2 cells). The color spectrum for the enrichment level (log2 scale) is the same as in Figure 5. The fraction of the heterochromatic repeats found in the BG3 extension regions is reported in the grayscale column on the right. The heterochromatic instances of all repeat types are marked by strong enrichment in HP1a, SU(VAR)3-9, and H3K9me2/3. In contrast, euchromatic repeat instances are associated with different types of chromatin patterns that vary in the levels of “active” and “silent” marks. Elements with similar patterns are marked by colored vertical bars on the left; red, highly enriched for “silent” marks, depleted for “active” marks; green, low enrichment or depletion for “silent” marks, highly enriched for “active” marks; orange, mixed enrichments for both “active” and “silent” marks. (B) Full-scale view of the top-most portion of the plot, showing repeat types for which euchromatic and heterochromatic instances show similar average chromatin patterns with predominant enrichments for “silent” marks. The RepBase repeat type names are shown on the left, with the number of instances found within each region to the right. In contrast, repeat types with mixed (C) and “active” (D) chromatin patterns in euchromatic regions (left) show predominantly “silent” mark enrichments when located in heterochromatin (right).











