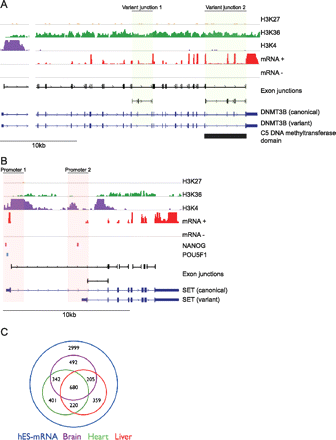
Alternative splicing in hES cells. (A) Integrated genome viewer (IGV) representation of the DNMT3B locus showing strand-specific RNA-seq data (mRNA±; scale: 0–5000 tags); exon junctions (black; detection threshold set at ≥8 tags); chromatin marks (H3K27, H3K36, and H3K4); two Ensembl transcripts (dark blue); and the C5 DNA methyltransferase functional domain (Pfam; bottom black). Detection of two non-canonical exon junctions (green shading) for DNMT3B indicated expression of at least two different transcript isoforms (dark blue), with expression of variant junction 2 predicted to result in truncation of the C5 DNA methyltransferase domain. (B) IGV view of the SET locus, including ChIP-seq data for POU5F1 (light blue) and NANOG (pink) binding sites, showing alternate promoter usage (red shading) and binding of both transcription factors at Promoter 1 and NANOG only at Promoter 2. (C) Venn diagram indicating tissue-specific expression of non-canonical alternative splicing events.











