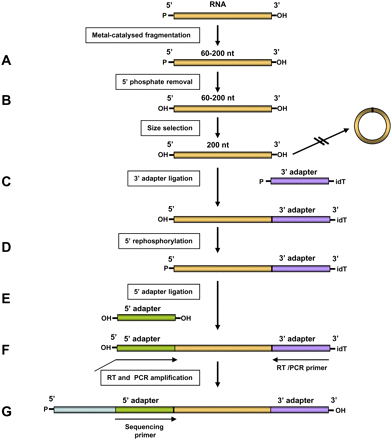
DSSS protocol workflow. (A) Fragmentation. RNA is fragmented to sizes in the range of 60–200 nt. (B) Dephosphorylation. 5′ phosphates are removed from RNA by treatment with alkaline phosphatase. (C) 3′ adapter ligation. Dephosphorylated 200-nt-long RNA fragments are selected by urea-PAGE. The 3′ adapter is ligated to the 3′ ends using T4 RNA ligase I. (D) Rephosphorylation. Fragments are rephosphorylated by treatment with T4 polynucleotide kinase as preparation for the next ligation step. (E) 5′ adapter ligation, preceded by removal of the nonligated 3′adapter by urea-PAGE size selection. (F) Reverse transcription (RT) and amplification of library. Molecules with 5′ and 3′ adapters were selected by urea-PAGE. First strand cDNA synthesis and PCR amplification were carried out with the indicated primers. (G) Sequencing.











