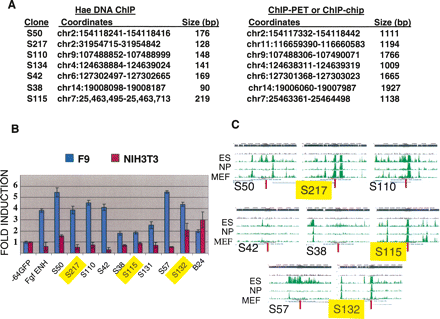
ChIP of NFRs followed by functional assays allows the high-resolution identification of functional target DNA sequences. (A) Overlap between functional SOX2-ChIP elements and previously reported target sequences. The genomic coordinates (assembly mm8) and the length of the DNA fragments for each of the target DNAs identified in each data set are indicated. (B) Transcriptional activation by the SOX2-ChIP’d NFRs listed in A, as well as several novel SOX2-ChIP HaeIII DNAs, were assessed after transfection of F9 or NIH3T3 cells. −64GFP and FgfENH are the negative and positive control plasmids, respectively (Fig. 1D), and GFP plasmids harboring each of the SOX2-ChIP’d NFRs are labeled as S. The control plasmid B24 (Supplemental Table 2) harbors a ubiquitously active HaeIII DNA. GFP expression elicited by each plasmid in F9 cells (blue bars) or NIH3T3 fibroblasts (red bars) were normalized to −64GFP and expressed as “fold induction.” (C) The genomic localization of each of the SOX2-ChIP elements (Supplemental Table 2) relative to H3K4me3 was determined as in Figure 3C. The elements highlighted in yellow correspond to proximal elements. The complete relationship of these elements with H3K4me3 is shown in Supplemental Figure 3.











