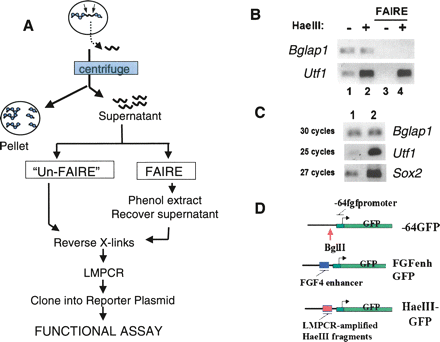
The HaeIII supernatants contain DNA derived from NFRs. (A) Method overview. Permeabilized nuclei are incubated with HaeIII. Silenced genes exhibit a densely packed nucleosome array (blue circles) that prohibits access of HaeIII to the DNA. This uncut DNA remains in the nuclei and is removed by centrifugation. In contrast, the relative absence of nucleosomes at the regulatory regions of actively transcribed genes renders the DNA accessible to HaeIII cleavage (small arrows). The released fragments diffuse out of the nucleus and can be recovered from the supernatant after centrifugation. (B) PCR analysis of HaeIII supernatants. F9 nuclei were incubated in the presence (+) or absence (−) of HaeIII. After centrifugation, DNAs in the supernatants were processed either with or without FAIRE treatment and analyzed for the relative levels of Bglap1 gene or Utf1 enhancer sequences using PCR. “Un-FAIRE” samples, lanes 1,2; FAIRE samples, lanes 3,4. (C) LMPCR enriches for DNA derived from NFRs. The relative levels of Utf1 enhancer, Sox2 enhancer, and Bglap1 DNA were compared before (lane 1) and after (lane 2) LMPCR amplification of DNAs in the HaeIII supernatants of the UnFAIRE sample. Note that after LMPCR, fewer cycles were required to visualize the Utf1 and Sox2 enhancer PCR products. (D) Reporter plasmids. The starting plasmid, −64GFP, contains the TATA box and transcription initiation site from the Fgf4 promoter cloned upstream of GFP coding sequences. The indicated BglII site was used for insertion of the Fgf4 enhancer to create the positive control plasmid FGFenhGFP, or LMPCR-amplified HaeIII fragments to create the HaeIII-GFP plasmid libraries.











