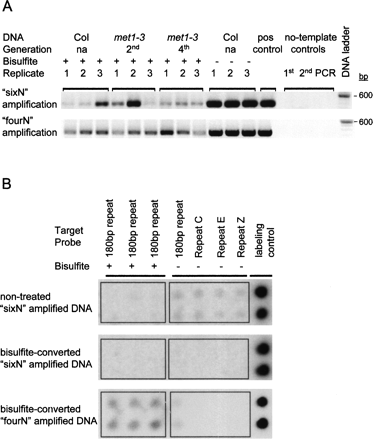
Post-amplification quality assessment following random amplification methods used for Bisulfite Methylation Profiling (BiMP). (A) Assessment for amplification bias using locus-specific PCR amplification at the AT3G08650 locus. For each DNA sample, two random amplification methods were performed, “sixN” and “fourN” (as indicated on the left) and assayed for the presence of the target template (see Supplemental Fig. 1A). The second- (2nd) and fourth-generation (4th) homozygotes of the met1-3 mutant are indicated. Col is indicated as “na” (not applicable). DNA sample treatment is indicated as either bisulfite-converted (+) or non-treated (−). Each lane corresponds to an equal volume of PCR reaction (10 μL). DNA was separated in a 1% agarose gel and stained with ethidium bromide. The PCR-positive control (pos control) was amplified from genomic Col DNA. The template-free controls (no-template control) correspond to the Sequenase elongation (1st), random amplification (2nd), and PCR (PCR) steps (see Methods). The gel images have been manipulated to create a contiguous lane order. (B) Evaluation of repeat representation in randomly amplified reaction products using dot blot hybridizations. Non-treated and bisulfite-converted Columbia DNA samples were amplified by the methods indicated (left) and hybridized to a macroarray (see Methods). The macroarray was spotted with 200 ng of PCR amplicons derived from Col DNA with (+) or without (−) bisulfite-conversion from different genomic repeat sequences (indicated above and described in the Methods). Each column of the macroarray was spotted with two technical replicates, observed as rows on the macroarray. Rectangular borders were added to the image to separate the bisulfite-converted (+) from the nonconverted (−) probes.











