Chromosomal rearrangement interferes with meiotic X chromosome inactivation
Abstract
Heterozygosity for certain mouse and human chromosomal rearrangements is characterized by the incomplete meiotic synapsis of rearranged chromosomes, by their colocalization with the XY body in primary spermatocytes, and by male-limited sterility. Previously, we argued that such X–autosomal associations could interfere with meiotic sex chromosome inactivation. Recently, supporting evidence has reported modifications of histones in rearranged chromosomes by a process called the meiotic silencing of unsynapsed chromatin (MSUC). Here, we report on the transcriptional down-regulation of genes within the unsynapsed region of the rearranged mouse chromosome 17, and on the subsequent disturbance of X chromosome inactivation. The partial transcriptional suppression of genes in the unsynapsed chromatin was most prominent prior to the mid-pachytene stage of primary spermatocytes. Later, during the mid-late pachytene, the rearranged autosomes colocalized with the XY body, and the X chromosome failed to undergo proper transcriptional silencing. Our findings provide direct evidence on the MSUC acting at the mRNA level, and implicate that autosomal asynapsis in meiosis may cause male sterility by interfering with meiotic sex chromosome inactivation.
Sex chromosomes exhibit unique behavior during the first meiotic prophase of spermatogenesis. Because they lack a homologous pairing partner, except for a tiny pseudoautosomal region (Perry et al. 2001), they undergo gradual condensation until the mid-pachytene stage, when the XY body is fully formed (Hoyer-Fender 2003). The condensed chromatin of the XY body is transcriptionally repressed by a process known as meiotic sex chromosome inactivation (MSCI) (Handel 2004; Turner 2007). The process begins in the late zygotene, when BRCA1-directed ATR kinase phosphorylates histone H2AX (Turner et al. 2004). MSCI is accompanied by additional variations in sex chromatin including the presence of dimethylated histone H3 on Lys9 (Khalil et al. 2004), heterochromatin protein 1 (HP1), histone macroH2A (Hoyer-Fender et al. 2000), ubiquitine ligase Rad18Sc (van der Laan et al. 2004), the high mobility group box protein MAEL (Costa et al. 2006); or the replacement of histone variants H3.1 and H3.2 by H3.3 (van der Heijden et al. 2007). The arrest of mouse spermatogenesis in prophase I is a frequent cause of male sterility due to mutations or deletions of functional genes, incompatibility of the parental genomes, or certain chromosomal rearrangements such as asymmetrical reciprocal translocations (Forejt 1996; de Rooij and de Boer 2003).
Previously, we proposed a possible link between the incomplete synapsis of chromosomes involved in male-sterile autosomal rearrangements and X chromosome inactivation in male meiosis (Forejt 1982; Forejt 1996). Here we investigated the phenomenon by analyzing both the aberrant chromosome synapsis with BRCA1 and gamma H2AX markers, and the global transcriptome changes with microarrays. Our studies revealed a significant transcriptional down-regulation of genes in the unsynapsed autosomal chromatin in pre-mid-pachytene spermatocytes that was followed by a partial breakdown of transcriptional silencing of the X chromosome during the mid-late-pachytene stage.
Results
Meiotic effects of T(16;17)43H translocation
To assess the meiotic effects of a male-sterile chromosomal rearrangement, we developed the C57BL/10-T43H/T43H congenic strain (hereafter B10-T43/T43) that carries the autosomal reciprocal translocation T(16;17)43H (hereafter T43) on the genetic background of the C57BL/10ScScPh (abbreviated B10) inbred strain. Consequently, by comparing B10-T43/T43, B10-T43/+, and B10-+/+ males, we could distinguish a possible position effect of the translocation break from the effect of asynapsis on the identical genetic background.
Only B10-T43/+ heterozygotes were completely sterile and displayed an incomplete synapsis of the translocation quadrivalent at the pachytene. The B10-T43/T43 translocation homozygotes and B10 males without translocation were fertile, had no pairing difficulties, and served as isogenic controls.
First, we investigated differences in the testicular cellularity of the mice of all three genotypes. Histological examination revealed severe reduction of spermatids and virtual absence of spermatozoa in seminiferous tubules of B10-T43/+ males when compared to fertile B10-T43/T43 and B10 controls (Supplemental Fig. 1). Fluorescence activated cell sorting (Bastos et al. 2005) enabled us to collect three major cell types: pre-mid-pachytene spermatocytes (comprising leptotene, zygotene, and early pachytene stages), the mid-late-pachytene cells, and spermatids. Sorting revealed an ∼12-fold lower ratio of mid-late-pachytene spermatocytes to pre-mid-pachytene spermatocytes in the B10-T43/+ heterozygotes than in the B10-T43/T43 homozygotes and B10 males. The ratio of spermatids to pre-mid-pachytene spermatocytes was even ∼19-fold lower, in agreement with the histological picture (Supplemental Fig. 1). Together, these findings confirmed a progressive spermatogenic breakdown at the pachytene level in heterozygous B10-T43/+ males.
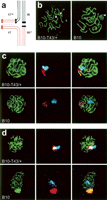
Next, we examined the synaptic status of chromosomes in individual pachytene spermatocytes. We anticipated pairing difficulties of rearranged autosomes of sterile B10-T43/+ males based on the extremely asymmetrical positions of the translocation breakpoints (Fig. 1a,b) and our previous studies (Forejt et al. 1981; Forejt 1984). The aberrant synapsis was, indeed, confirmed by the immunostaining of BRCA1 and gamma H2AX (Fig. 1c,d), the markers of the unsynapsed chromatin and the XY body (Scully et al. 1997; Turner et al. 2004). Immunostaining of the BRCA1 revealed a partial asynapsis of translocated autosomes in 61% (31/51) of the pachytene spermatocytes from sterile males (Fig. 1c). Moreover, gamma H2AX appeared in 70% (37/53) of pachytene spermatocytes either as enlarged or germinated foci in close contact with the T43 translocation quadrivalent (Fig. 1d). This result resembles the BRCA1 pattern. Under normal conditions, the X and Y chromosomes are transcriptionally silenced by MSCI and separated from the active chromatin in the XY body. However, the colocalization of translocated autosomes and sex chromosomes accompanied by an abnormal distribution of gamma H2AX in pachytene spermatocytes of B10-T43H/+ males pointed to an abnormal formation of the XY body. Based on this and earlier studies (Forejt et al. 1981; Sciurano et al. 2007), we suggest that the intimate contact of unsynapsed autosomal segments with XY chromatin discloses the time and space of their position-effect-variegation-like interactions.
Synapsis and XY body formation in pachytene spermatocytes of B10-T43/+ sterile males and B10 controls. (a) Scheme of pachytene synapsis in the translocation quadrivalent of B10-T43/+ males. Proximal (centromeric) parts of chromosomes 17 and 1716 are most prone to synaptic failure. (b) Synaptonemal complexes were labeled with anti-SCP3 antibody. The arrowheads indicate the axial elements of sex chromosomes; the arrow points to the aberrantly synapsed translocated autosomes. (c) The unsynapsed chromatin was detected by immunostaining with anti-BRCA1 antibody (red). BRCA1 was immunolocalized on the sex chromosomes of sterile and fertile males and within translocated chromosomes in the sterile male. The SCP3 visualized axial elements (green); chromosome painting identified chromosome 17 (light blue). (d) Spread spermatocytes from sterile and fertile males were coimmunostained with anti-SCP3 (green) and anti-gamma H2AX (red) to disclose differences in XY body formation. Painted chromosome 17 is shown in light blue.
Transcriptional down-regulation of unpaired autosomal chromatin in pre-mid-pachytene spermatocytes
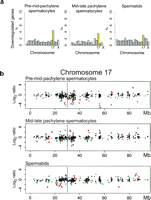
To evaluate the transcriptional behavior of individual chromosomes in the sterile B10-T43/+ males and their fertile congenic B10-T43/T43 and B10 counterparts, we compared their transcriptomes in sorted populations of pre-mid-pachytene spermatocytes, mid-late-pachytene spermatocytes, and spermatids. Since the genetic background of analyzed animals was virtually identical except for the presence of the T43 translocation, any expression differences between sterile and fertile males should relate to chromosomal rearrangement. In the pre-mid-pachytene spermatocytes of B10-T43/+ mice, 4.48% (35/782) of all chromosome 17 genes with a probe set on the Affymetrix MG_430 2.0 GeneChip were down-regulated, equal to 9.4% (35/373) of expressed genes. The abundance of the down-regulated genes was significant (P ≤ 0.0018; one-tailed Fisher exact tests) when compared to any other chromosome (Fig. 2a). The phenomenon was still significant in the mid-late-pachytene spermatocytes when we compared chromosome 17 to any other chromosome (P ≤ 0.017), except for chromosome 15 (P = 0.065).
Down-regulation of genes in the vicinity of the translocation breakpoint. (a) Percentage of down-regulated genes in individual chromosomes of sterile B10-T43/+ males compared to fertile controls (B10 and B10-T43/T43). Significant enrichment of down-regulated genes (P < 0.05) was found on chromosome 17 (yellow) in pre-mid-pachytene spermatocytes (P ≤ 0.0018; one-tailed Fisher exact tests). (b) Expression changes between sterile and fertile males along chromosome 17 are shown in a logarithmic scale of base 2. These changes were calculated as [(BT/B + BT/T)/2], where BT, B, and T stand for the mean expression in B10-T43/+, B10, and B10-T43/T43 males, respectively. Significant expression differences are plotted in green (P < 0.05) and red (P < 0.01). Only the genes expressed in the specified cell population from males of at least one genotype are shown. (Blue line) The translocation breakpoint, localized at ∼30 Mb.
The distribution of the down-regulated genes along chromosome 17 was highlighted on the chromosome map as expression ratios between sterile and fertile males (Fig. 2b). The down-regulated genes were not distributed randomly. They clustered in an interval of maximum asynapsis of the T43/+ translocation quadrivalent around 30 Mb, in the vicinity of the T43 translocation breakpoint (Vacik et al. 2005). We compared the observed proportion of down-regulated genes to the proportion obtained by 100,000 random permutations of the gene order on the entire chromosome 17 in order to test the enrichment of down-regulated genes in the chromosomal interval spanning the translocation breakpoint (20 Mb to 40 Mb). Significant enrichment was observed only in the pre-mid-pachytene spermatocytes (P = 0.00078), where 14.5% (26/179) of the expressed genes in this region were down-regulated. In the later stages of spermatogenesis, the proportion of down-regulated genes decreased to 10.2% (17/166) in mid-late-pachytene spermatocytes and 6.6% (11/167) in spermatids, respectively. The observed gene down-regulation in the area of asynapsis provides the first direct evidence that meiotic silencing of unsynapsed chromatin (MSUC) (Schimenti 2005; Turner et al. 2005) operates at the mRNA level. The down-regulation of five selected genes of chromosome 17 was confirmed by real-time quantitative RT-PCR (Supplemental Fig. 2).
Incomplete silencing of the X chromosome genes in mid-late-pachytene spermatocytes
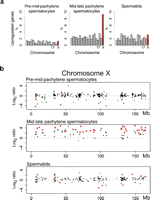
Next, we analyzed the genes overexpressed in the sterile B10-T43/+ males by comparing them to B10 and B10-T43/T43 isogenic controls (Fig. 3a). At the mid-late-pachytene stage, 9.3% (53/571) of all X chromosome genes with probe sets on the GeneChip were up-regulated in B10-T43/+ males when compared to the control fertile males. This is a significantly higher frequency than the frequency of up-regulated genes on any autosome (P ≤ 0.00017; one-tailed Fisher exact tests), and it corresponds to 65.4% (53/81) of the expressed X-linked genes. As 224 genes from the X chromosome were expressed in the pre-mid-pachytene spermatocytes, the improper silencing in sterile males was found for 23.7% (53/224) of X-linked genes expressed prior to MSCI.
Overexpression of genes on the X chromosome in mid-late pachytene spermatocytes of sterile males. (a) Percentage of overexpressed genes in individual chromosomes of sterile B10-T43/+ males compared to fertile controls (B10 and B10-T43/T43). Significant enrichment of overexpressed genes (P < 0.05) was found on chromosome X (red) at mid-late pachytene (P ≤ 0.00017; one-tailed Fisher exact tests). (b) Distribution of expression changes between the sterile and fertile males along the X chromosome. These changes were calculated as in Figure 2. Significant expression differences are plotted in green (P < 0.05) and red (P < 0.01). Only the genes expressed in the specified cell population from males of at least one genotype are shown.
The distribution of the up-regulated genes was uniform along the X chromosome (Fig. 3b) in contrast to the clustering of down-regulated genes observed on chromosome 17. The overexpression of six X-linked genes in sterile males was confirmed by real-time quantitative RT-PCR (Supplemental Fig. 2). The relative overexpression of the X chromosome genes was not apparent either in pre-mid-pachytene spermatocytes or in the spermatids of sterile males.
Finally, we assessed the rate of the X chromosome expression variations by calculating the global X:autosome expression ratio (Nguyen and Disteche 2006) known to reflect the course of MSCI. As expected, the mid-late-pachytene spermatocytes from B10-T43/T43 and from B10 fertile males displayed an extremely low X:autosome ratio (0.08) because of the effective silencing of the X-linked genes via MSCI. A much higher ratio (0.37) in the B10-T43/+ sterile males confirmed the partial failure of meiotic silencing (Fig. 4).
Relaxed silencing of the X chromosome reflected by a high X:autosome transcription ratio in sterile B10-T43/+ males. The X-to-autosome transcription ratios are shown for individual cell populations and mouse genotypes. For each genotype, the mean ratios of two independently isolated populations are displayed together with their range. BT, B, and T stand for B10-T43/+, B10, and B10-T43H/T43H genotypes, respectively.
Discussion
We analyzed the meiotic effects of mouse T(16;17)43H translocation, our model of a male-sterile chromosomal rearrangement. We have obtained direct evidence on the transcriptional silencing of unsynapsed autosomal chromatin (MSUC) of the translocation quadrivalent and on the subsequent failure of proper transcriptional inactivation of the X chromosome (MSCI) at the mid-late pachytene. The colocalization of rearranged autosomes and XY chromosomes at pachytene suggested MSUC as a cause of disturbed MSCI.
The colocalization of the XY bivalent and rearranged autosomes was discovered in mice (de Boer and Groen 1974; Forejt 1974). Later on, its further occurrence was confirmed in mice (de Boer and Branje 1979; Forejt and Gregorova 1977; Forejt 1979, 1996; Forejt et al. 1981), in humans (Johannisson et al. 1983; Gabriel-Robez and Rumpler 1996; de Boer et al. 2004; Oliver-Bonet et al. 2005; Sciurano et al. 2007), and in other mammals (Power 1991; Jaafar et al. 1992; Ansari et al. 1993). These cytogenetic studies revealed the coincidence of chromosomal asynapsis, XY-autosomal colocalization, and male-limited sterility. However, the evidence was not sufficient to verify the idea of disturbed MSCI as a common cause of spermatogenic arrest in carriers of otherwise unrelated chromosomal rearrangements. More recently, the phenomenon of meiotic silencing of unpaired DNA was uncovered in Neurospora (Shiu et al. 2001) and Caenorhabditis elegans (Bean et al. 2004). In the mouse, MSUC was identified in unsynapsed parts of rearranged chromosomes involved in T(X;16)16H translocation (Turner et al. 2004, 2005) and in double heterozygotes for an overlapping translocation, T(1;13)70H/T(1;13)1Wa (Baarends et al. 2005; van der Heijden et al. 2007). These studies showed that an unpaired chromatin, much like the XY pair, displayed BRCA1, ATR, and gamma H2AX, and showed an exclusion or reduction of Cot-1 painting, both indications of transcriptional silencing. It has to be stressed that this manifestation of MSUC was found in male as well as female meiotic prophase, suggesting that MSUC per se cannot explain the male-specific arrest of gametogenesis.
The epigenetic modifications of male and female unsynapsed chromatin further included the ubiquitination of H2A histone, the H3K27 trimethylation, and the dimethylation of histone H3 at lysine-9. High mobility group protein MAELSTROM and interacting SIN3B protein were also found on the XY body and in unsynapsed autosomes (Costa et al. 2006). Recently, an indication that MSUC can interfere with MSCI has been observed in pachytene spermatocytes where H3.1 and H3.2 histones of the sex chromosomes are replaced by the H3.3 variant. In T(1;13)70H/T(1;13)1Wa double heterozygotes, this nucleosome replacement occurred within the unsynapsed autosomal chromatin but failed to proceed to completion in adjacent sex chromosomes, thus indicating their disturbed MSCI (van der Heijden et al. 2007).
Our analysis of T43/+ translocation confirmed the localization of BRCA1 and gamma H2AX on the unsynapsed arms of the translocation quadrivalent. We have shown that a significant fraction of the genes within the unsynapsed chromatin is transcriptionally down-regulated and that the transcriptional silencing of the X-linked genes is disturbed. The incomplete transcriptional silencing of genes within the unsynapsed interval of chromosome 17 can reflect cell-to-cell heterogeneity of asynapsis and/or the differential sensitivity of individual genes to silencing as known from classical studies on position effect variegation. Rather unexpectedly, the transcriptional silencing within the unsynapsed autosome was most prominent early in meiotic prophase, in the fraction of pre-mid-pachytene spermatocytes, and before the disturbance of X-inactivation became apparent. The disturbance of X-inactivation was most obvious during mid-late pachytene, when autosomal silencing had already been decreasing. Turner and colleagues described a wave of H2AX phosphorylation targeted to XY and unsynapsed chromatin already during zygotene (Turner et al. 2005). The fact that they detected transcriptional silencing at pachytene, while we could see it already at zygotene and early pachytene, could be explained by different sensitivity of the methods used (Cot-1 FISH vs. microarray expression profiling). Based on our current knowledge and on the global transcriptome analysis of the T43 autosomal translocation, we can update our previous hypothesis (Forejt 1984, 1996) on the mechanism of spermatogenic arrest in carriers of male-sterile rearrangements into the following consecutive steps: (1) The chromosomal rearrangement prevents the full extent of the homologous synapsis of the chromosomes involved. (2) The unsynapsed parts of the rearranged chromosome(s) recruit specific proteins, including BRCA1, ATR, and gamma H2AX, engaged in meiotic silencing of unsynapsed chromatin. (3) The modified autosomal chromatin is attracted, by a yet unknown mechanism, to intimate contact with the XY chromosomes. (4) The partially silenced autosomal chromatin interferes with MSCI and causes incomplete histone substitution (de Rooij and de Boer 2003) and disturbed transcriptional inactivation (present data) of the sex chromosomes. (5) The interference with MSCI functions as a trigger of apoptosis and results in spermatogenic arrest.
A corollary to the male-limited character of sterility induced by meiotic silencing of unsynapsed chromatin is the fact that although MSUC operates in both sexes, it can interfere with the X chromosome inactivation only in male meiotic prophase.
Methods
Mice
The effect of the T(16;17)43H (Searle et al. 1974) (T43) chromosomal translocation on spermatogenesis was studied in congenic males that were genetically identical with the exception of the translocation break and its flanking sequences. To construct the B10-T43/T43 congenic mice, we repeatedly backcrossed T43 translocation to the C57BL/10ScSnPh (B10) inbred strain and made it homozygous through a fertile intermediate, a B10-T43/B10-Rb7(16;17)Bnr double heterozygous male (Forejt et al. 1995). All male mice used in the present experiments were 2 mo old.
The mice were kept at the Specific Pathogen-Free Facility of the Institute of Molecular Genetics, AS CR. The principles of laboratory animal care obeyed the Czech Republic Act for Experimental Work with Animals (Decrees No. 207/2004 Sb., and the Laws Nos. 246/92 Sb. and 77/2004 Sb.), fully compatible with the corresponding EU regulations and standards, namely, Council Directive 806/609/EEC and Appendix A of the Council of Europe Convention ETS123.
Immunostaining of spread spermatocytes and chromosome painting
Meiotic cell spreads were prepared as described (Turner et al. 2005) with modifications. The spermatogenic cells were treated with CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2 10 mM PIPES at pH 6.8, 0.5% Triton X-100, 0.5 mM EDTA, 1× protease inhibitors [Roche]) for 10 min, fixed by 2% paraformaldehyde (Electron Microscopy Sciences), and rinsed in PBS. The slides were blocked by PBT (PBS, 0.5% BSA, 0.1% Tween 20) and incubated with primary antibodies diluted in PBT in a humid chamber overnight at 4°C. Rabbit polyclonal anti-SCP3 antibody (Lammers et al. 1994) was used at 1:1000 dilution. Primary mouse monoclonal antibody against gamma H2AX (Upstate, cat. no. 05-636) was used at 1:2000 dilution, and antibody against BRCA1 (GH118) (see Ganesan et al. 2002) was used at 1:3 of hybridoma supernatant. Slides were washed three times for 5 min in PBS, followed by incubation with secondary antibodies diluted in PBT for 2 h at 4°C. The secondary antibodies were goat anti-rabbit FITC (Sigma-Aldrich) at 1:100 and goat anti-mouse Alexa 594 (Molecular Probes) at 1:500. Slides were washed three times for 5 min in PBS, mounted, and counterstained by a mounting medium with DAPI (Vector, UK). Fluorescence in situ hybridization (FISH) was performed with mouse whole-chromosome paint probes after immunofluorescence microscopy according to the manufacturer’s instructions (Cambio). Chromosomes were painted using FITC-labeled mouse chromosome 17 and/or the Cy3-labeled mouse X chromosome. Images were captured in a Nikon Eclipse 400 microscope using a Pixera Penguin 600CL cooled CCD camera (Pixera). The captured images were superimposed and background subtracted, and the original green signal of chromosome 17 was converted to a light blue color with Adobe Photoshop (Adobe Systems).
Histology
Testes of mice were dissected and fixed with 10% paraformaldehyde in phosphate-buffered saline overnight. The fixed testes were embedded in paraffin, sectioned at 2–3-μm thickness, and mounted on slides. The mounted sections were deparaffinized, rehydrated, and processed for standard histological stains (Mason’s blue trichrome and hematoxylin/eosin). The specimens were observed on a Nikon Eclipse E800 microscope. The images were captured using a DP70 CCD camera (Olympus) and processed with Adobe Photoshop (Adobe Systems).
FACS characterization and isolation of spermatogenic populations
Populations of pre-mid-pachytene spermatocytes, mid-late-pachytene spermatocytes, and spermatids were isolated using fluorescence-activated cell sorting according to Bastos et al. (2005) with minor modifications. The cell populations were sorted in duplicates, from two animals of each of the three genotypes: B10-T43H/+, B10-T43H/T43H, and B10. Briefly, spermatogenic tubules of mice euthanized by cervical dislocation were incubated in enriched Krebs-Ringer bicarbonate medium (EKRB) with collagenase (0.5 mg/mL; Sigma) for 20 min at 32°C on a shaker. The tubules were filtrated with a cell strainer (BD Falcon) and incubated with collagenase under the same conditions. The suspension was filtered, and the cells were washed twice by EKRB containing 1% FCS. Finally, the cells were diluted in 1 mL of EKRB with 1% FCS and stained with Hoechst 33342 (5 μg/mL) for 1 h at 32°C. Propidium iodide was added just before FACS analysis to a concentration of 2 μg/mL. Individual populations were sorted according to red and blue Hoechst emission (Bastos et al. 2005) directly into the RLT buffer of the RNeasy micro isolation kit (QIAGEN). Aliquots were sorted to EKRB medium for subsequent immunofluorescence analysis. Based on SCP3, SCP1, and gamma H2AX immunostaining and morphology of the cells, we have determined the composition of FACS-isolated populations. The fraction of pre-mid-pachytene spermatocytes consisted of leptotene and zygotene spermatocytes (∼60%), and early-pachytene spermatocytes (∼40%). The population of mid-late-pachytene spermatocytes displayed ∼90% of the cells at the late-pachytene stage. The population of spermatids consisted of ∼90% of the spermatids.
Microarray analysis
The total RNA (15–40 ng) was converted in cRNA using the Affymetrix Two-Cycle Target Labeling kit according to the manufacturer’s instructions. Eighteen Affymetrix GeneChip Mouse Genome 430 2.0 Arrays were hybridized with cRNA obtained from the isolated populations. The data were analyzed with Bioconductor 1.9 (Gentleman et al. 2004; http://www.bioconductor.org/) and the R project for statistical computing (version 2.3; http://www.r-project.org/). The probes were annotated to Entrez gene identifiers (NCBI build 36) using the custom chip description file (cdf) “mm430mmentrezg8” (Dai et al. 2005), from the Bioconductor repository. The data were normalized using gcRMA (Irizarry et al. 2003), and the probe sets with unknown chromosome localization were filtered out. Only genes having a corresponding probe set intensity >50 in at least one of the three genotypes studied were considered expressed in the particular cell population.
Statistics
We used Linear Models for Microarray Data Package, limma version 2.9.10 (Smyth 2005) for statistical evaluations of expression differences. A linear model was fitted for each gene in a given series of arrays by using the lmFit function. To rank the differential expression of genes, we applied the eBayes function of the empirical Bayes method. A correction for multiple testing was performed using the Benjamini and Hochberg false-discovery-rate (FDR) method (Benjamini and Hochberg 1995). We considered genes to be differentially expressed if the adjusted P-value was <0.05 for each of the two comparisons: B10/T43H versus T43H/T43H, and B10/T43H versus B10/B10, and exceeded the threshold value 50 in at least one of the mouse genotypes.
To assess the significance of enrichment of down-regulated genes on chromosome 17, we compared their incidence on chromosome 17 and on other chromosomes by a series of one-tailed Fisher exact tests. Chromosome Y was omitted because only eight genes mapped to the probe sets. The obtained P-values were adjusted using the Benjamini and Hochberg FDR method to correct for multiple testing. The same approach was used to assess the significant incidence of up-regulated genes on chromosome X.
To learn if the down-regulated genes tend to cluster within the chromosome 17 interval encompassing the translocation breakpoint (sequence coordinates: 20–40 Mb), we performed 100,000 permutations of the order of the genes expressed from this chromosome. The P-value (one-tailed) was computed as a number of permutations yielding counts of significantly down-regulated genes in the specified area of chromosome 17 above or equal to the observed gene counts.
The X:autosome expression ratio was calculated for each cell population and mouse genotype as the fold difference of mean probe set intensities of the X-linked versus autosomal genes.
Real-time quantitative RT-PCR
Total RNA isolated from spermatogenic populations was reverse-transcribed using Mu-MLV Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed with the Light-Cycler DNA Fast Start Master SYBR green I kit (Roche) on a Light Cycler 2000 machine at Tm = 61°C. The sequences of primers are in Supplemental Table 1.
Acknowledgments
We thank C. Heyting, S. Dimitrov, and D.M. Livingston for providing antibody reagents; Z. Cimburek for help with FACS analysis; J. Sikora for histological sections; and P. Divina, Z. Trachtulec, and S. Takacova for critically reading the manuscript. This work was supported by grants nos. 301/06/1334 and 301/07/1383 from The Czech Science Foundation and nos. IM6837805002 and AV0Z50520514 from the Ministry of Education, Youth and Sports of the Czech Republic. J.F. was supported as an HHMI International Research Scholar.
Footnotes
-
↵1 Corresponding author.
↵1 E-mail jforejt{at}biomed.cas.cz; fax 420-24106-2154.
-
[Supplemental material is available online at www.genome.org. The microarray data have been submitted to NCBI/GEO under accession no. GSE7306.]
-
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6520107
-
- Received March 20, 2007.
- Accepted July 3, 2007.
- Copyright © 2007, Cold Spring Harbor Laboratory Press