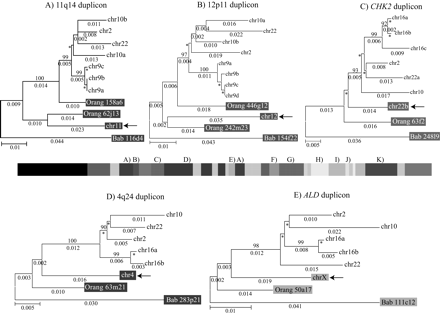
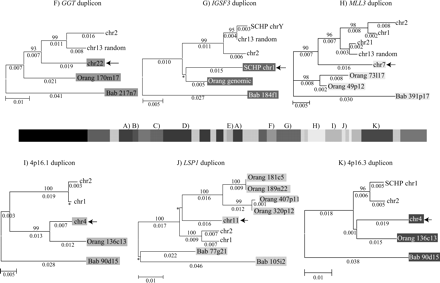
Phylogenetic trees for 2p11 duplicons. A neighbor-joining tree was constructed for each individual duplicon as shown above and below the gray schematic of the 2p11 duplicons (A–K). See Figure 1 for corresponding colored boxes. Gray boxes outline ancestral human, orangutan (Orang), and baboon (Bab) sequence taxa within the phylogenetic trees. Ancestral human sequences are also marked with an arrow. Branch lengths are proportional to the number of nucleotide changes between taxa and are indicated below each respective branch. An asterisk next to or below a branch length indicates a branch length of 0.001. Bootstrap values >90 from 1000 replicates are indicated above each corresponding branch. Sequence data from baboon and orangutan outgroups were obtained from large-insert BAC clones (CHORI-253 and RPCI-41) or total genomic DNA.











