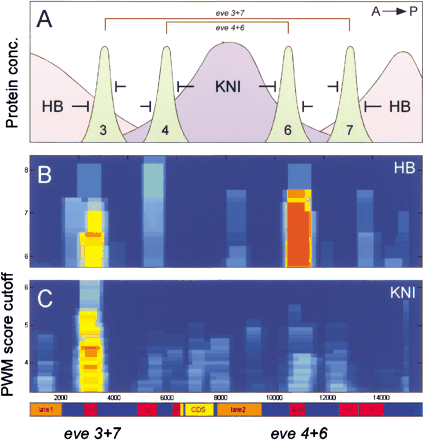
Quantitative description of gene response to TFs. Overlapping Hunchback and Knirps gradients combine two symmetrical zones, where theeven-skipped expression stripes 4+6 and 3+7 are formed (A). The Hunchback cluster in the eve 3+7 enhancer has a lower PWM cutoff (site affinity) and a lower statistical significance (less sites) than that in the eve 4+6. In contrast, the Knirps cluster in eve 3+7 enhancer has more high-affinity sites than that in the eve 4+6 (B). The detected composition of clusters for Hunchback and Knirps in theeven-skipped locus supports the existing biological model and might be used for interpretation of gene response to the upstream regulators.











