Time for a Unified System of Mutation Description and Reporting: A Review of Locus-Specific Mutation Databases
- 1Genomic Disorders Research Centre, St. Vincent's Hospital Melbourne, Fitzroy VIC 3065, Australia, 2Laboratoire de Génétique Moléculaire, Institut Universitaire de Recherche Clinique, 34093 Montpellier Cedex 5, France, 3The University of Melbourne, Department of Medicine, Melbourne VIC 3010, Australia
Abstract
Mutation databases of human genes are assuming an increasing importance in all areas of health care. In addition, more and more experts in the mutations and diseases of particular genes are curating published and unpublished mutations in locus-specific databases (LSDB). These databases contain such extensive information that they have become known as knowledge bases. We analyzed these databases and their content between June 21, 2001, and July 18, 2001. We were able to access 94 independent websites devoted to the documentation of mutation containing 262 LSDBs for study. We analyzed one LSDB from each of these websites (i.e., 94 LSDBs) for the presence or absence of 80 content criteria, as generally each gene in a multigene website documented the same criteria. No criterion studied gave unanimous agreement in every database. Twenty-two genes were represented by more than one LSDB. The number of mutations recorded, excluding p53, was 23,822 with 1518 polymorphisms. Fifty-four percent of the LSDBs studied were easy to use and 11% hard to follow; 73% of the databases were displayed through HTML. Three databases were found that were given a high score for ease of use and wealth of content. Thus, the study provided a strong case for uniformity of data to make the content maximally useful. In this direction, a hypothetical content for an ideal LSDB was derived. We also derived a community structure that would enhance the chances of mutation capture rather than being left unpublished in a patient's report. We hope the interested community and granting bodies will assist in achieving the vision of a public system that collects and displays all variants discovered.
[Supplemental material available online at http://www.genome.org]
Both completion of the human genome sequencing project and new methods for the detection of point mutations, such as by microarray chips, will lead to a tremendous increase of mutation identification in a growing number of genes. Consequently, the task of reporting and analyzing germline or somatic DNA variation will be a major challenge for the future of biological and medical science. Mutation databases are repositories in which allelic variations are described and assigned within a specific gene. Currently, two types of databases are available: central databases and locus-specific databases (LSDBs) (Horaitis and Cotton 1999). Genome-wide general or central mutation databases contain pooled information on variation across the whole genome and have developed tools for analyzing existing data collections while providing consistent user interfaces to all genes. LSDBs concentrate on variation within a single gene and are usually run by a consortium of collaborating researchers with scientific expertise in a particular gene or phenotype. Some curators are responsible for a number of LSDBs at a single site. None of the central databases are sufficient themselves for the needs in medical genetics; they are a complementary resource to LSDBs and each benefits from the presence of the other. Central mutation databases and some LSDBs were recently described in a special issue of Human Mutation (2000). The Human Genome Organization (HUGO) Mutation Database Initiative (MDI) recently formed into the Human Genome Variation Society (HGVS) and maintains a dedicated website documenting a catalog of LSDBs and the directory of curators; it is ever growing and has been described in print also (Horaitis et al. 2001).
LSDBs provide an invaluable tool for analyzing gene expression and phenotype in both normal and disease conditions, as the curators are closely in touch with molecular biologists very experienced with the analysis of a specific gene and its anomalies. This system generally promotes submission of data and maintains an accurate and up-to-date data source. LSDBs were developed independently of other analogous databases, so they have different content and structure depending on gene and disease characteristics. It is essential, however, that they share a minimum of core elements to be usable for the majority of the community. These include the following: a unique identifierfor the allele; the source of the data (published article, abstract, investigator); the context of the allele (species, gene, reference sequence); and the allele itself (name, type, nucleotide change, and so forth) (Scriver et al. 1999, 2000).
To better document the diversity of existing LSDBs and provide a guide to future activity, we have analyzed the structure and content of each of the LSDBs currently available through the World Wide Web (WWW).
RESULTS
We examined a total of 94 independent websites maintained by curators in 17 countries that describe mutations associated with human disease, including 65 sites with one LSDB and 29 multigene sites (Table1A). Of 262 LSDBs available to our knowledge, 30 were redundant, leaving 232 different nuclear genes with an LSDB. By way of comparison, the Human Gene Mutation Database (HGMD) (Krawczak et al. 2000) lists 1044 genes that contain at least one mutation. The redundancy found among LSDBs indicated that mutations were reported by two databases in the case of 18 genes, three databases in the case of three genes, and six different databases reported the gene TP53 (Table 1B). A total of 23,822 mutations and 1,518 polymorphisms were recorded on July 16, 2001. They were mostly different mutations in each gene; however, because of redundancy of databases and lack of standardization of mutation entries, it was impossible to know what is the nonredundant set of mutations entered into available LSDBs. The number of mutation records was 53,715 if TP53 variation was included. Raw data of the analysis can be seen in Table A at www.genome.org and a summary in Table 2. Further inquiries regarding data and methods can be directed to the authors.
Current Status of Locus-Specific Databases (LSDBs)
Summary of Raw Data for LSDBs
Criteria Examined
General Presentation of LSDBs
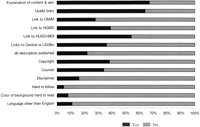
Eighteen percent of LSDBs originated in a consortium (the number of investigators ranged from 2 to 138) with shared interests in annotating allelic variants. Most LSDBs had a home page that provided a clear explanation of content and aim of database and a minimum set of cross-references (active links and pointers) for the user to access additional information (Fig. 1). Important links included HGMD and other central mutation databases for information on genetic variation, OMIM (Online Mendelian Inheritance in Man) for clinically related information, MedLine/PubMed for access to published references, GenBank/EMBL/DDBJ for detailed DNA sequence information, HUGO nomenclature database, and other useful links. Thirty eight percent of LSDBs advised users that information in the database is copyrighted intellectual property, such that they should cite the database in the appropriate manner when using data. This study also reveals that only 54% of LSDBs would fit minimal criteria for both an easy and optimal use of the information that they contain. Curiously, the color of the background made reading hard in 8% of the databases. Overall, we found that data in 11% of LSDBs were hard to follow.
Data Collection and Submission (Data Source)
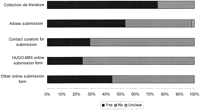
LSDBs were composed of mutation entries, such that each entry usually corresponds to a mutation in a single patient and is added (generally, but not always) after curator inspection (Fig.2). Most databases were a compilation of data derived from both published literature (75%) and submissions directly to the LSDB contributed by researchers throughout the world (53%). Data were submitted directly to curators by filling in an online questionnaire in 68% of LSDBs that allowed submission. Of these, 24% used the specially designed HUGO-MDI entry form. Submission by contacting curators directly was available in 29% of cases.
Information on Disease, Gene, and Protein
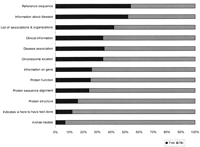
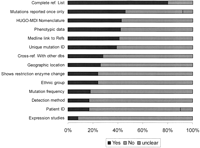
A number of databases contained much information in addition to the list of mutations, making the registries valuable for physicians and scientists from many fields. About half of the LSDBs provided information on the disease and/or the gene associated with the mutations that they described, whereas other LSDBs had just a link to OMIM or other sites for clinical information or showed only a list of mutations (Fig. 3). Information on the gene of interest, protein function, protein structure, and protein sequence alignment could be found in some LSDBs. A few of the LSDBs function as “knowledge bases” because they combine scientific and diagnostic data on mutations with associated information useful for clinicians or students (e.g., population distribution of alleles, haplotype associations) and information for patients and their families (e.g., treatment, diagnosis, dedicated organizations, or parent associations). Some LSDBs aimed to facilitate the detection and characterization of mutations by providing technical support in the form of primer sequences and mutation detection protocols.
Mutation Database Structure and Software
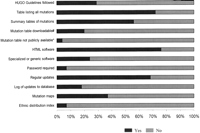
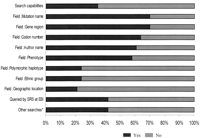
Access to mutation information was usually free; a few of the databases (7%) were password protected and registration for membership was requested to ensure that individuals using the database agree to a set of guidelines covering data submission, confidentiality, appropriate data use, and acknowledgment (Fig.4). We found that 4% of LSDBs were not publicly available, including one subscription-based access database. Mutation tables were structured mostly as flat files containing a number of fields for each entry. A complete table listing all mutations was available in only 72% of LSDBs, including downloadable formats sometimes difficult or impossible to download. Summary sheets describing the total number of alterations reported in each exon or the number of these variants that are distinct or ethnic distribution formats were also available in a number of LSDBs. Some databases showed mutation maps depicting the location of mutations throughout the gene (or even the protein) sequence, and a few added graphical displays, including dynamic graphing tools. HUGO-MDI guidelines (Scriver et al. 1999, 2000) for the construction of database were followed in 29% of LSDBs.
Up to 73% of LSDBs were displayed on the WWW through Hypertext Markup Language (HTML). Because there is no standard yet, the way data is presented in LSDBs varies from simple flat-file-type databases, which are listings of the mutations in the specific gene plus their publication references, to fully interactive databases that can present data in a multitude of ways. Most curators used flat file, plain text databases or spreadsheet programs (such as MicrosoftExcel) as a simple means to collate and store data on mutations. Only 27% of curators used specialized or generic software such as the Universal Mutation Database (Beroud et al. 2000), the Mutation Storage and Retrieval Program (MuStaR)(Brown and McKie 2000), or other programs.
Mutation Table and Mutation Documentation
The mutation table (found in 72% of LSDBs) listed all types of mutations stored in the database (Fig. 5). Recommended HUGO-MDI nomenclature (Antonarakis and the Nomenclature Working Group 1998) was used by 42% of LSDBs. A unique identifier (ID) number for each mutation record was found in 39% of LSDBs, and 17% had a patient ID. One site proposed an interesting patient identity number (PIN) that names the patient and the mutation in an unambiguous manner. In addition to the mutation listing, many databases provided data fields for associated information, such as mutation detection methodology (17%), mutation frequency (18%), ethnicity (24%) or geographic origin of patients (26%), restriction enzyme change with mutation (24%), and expression studies (8%), as well as information about the frequency of specific variants. Mutations were reported only once in 46% of LSDBs. A complete reference list of authors who reported the mutation was shown in 80% of databases, with a direct link to Medline/PubMed ID in 41%.
Querying the Database
A search engine in 35% of databases provided the opportunity to interrogate the database for specific information contained in a number of fields (gene symbol, mutation type, intragenic position, nucleotide change, amino acid change, restriction enzyme change, CpG hot spot, population, geographic location, phenotype, reference, and so forth) (Fig. 6). More complex queries could also be constructed by 41% of websites using the Sequence Retrieval System (SRS), a cross-link program maintained at the European Bioinformatics Institute (Lehväslaiho et al. 1998;2000), which had mounted information from the 94 websites in 41 cases.
Scoring LSDBs for Ease-of-Use and Information Content
Arbitrary scores (scale 1 to 10) were assigned by one of us, inexpert with mutation databases, for ease of access and browsing, as well as for evaluating quantity and quality of information contained in LSDBs (Table 3). We found that 48 of 94 scorable databases (51%) were >5, and three (3.1%) had a maximal score of 9; no LSDB reached the ideal score of 10. The top three scoring databases were the Phenylalanine Hydroxylase Knowledge Base, the Human Haemoglobin Variants home page, and the Blood Group Antigens home page. It is possible other users may rank them very differently.
Scoring of Locus-Specific Databases
Comparison of LSDBs with Central Databases
We analyzed the data for a particular gene (PAH) in two central databases, HGMD and OMIM, to define the differences between the content of LSDBs and such databases (See Supplementary Table A atwww.genome.org). The most striking absence in the central databases was the lack of the ability for searching the database. OMIM lists a limited number of mutations and HGMD contains limited gene information (as did many LSDBs), for example, no disease association, frequency, methods, ethnicity, geographic origin, or list of associations. Although MITOMAP, a human mitochondrial genome database was also surveyed, it may be regarded as a hybrid between an LSDB and a central database, its fields are more similar to an LSDB with a notable searching facility.
The Keio database was not viewable in the study period as a result of password restrictions; however, this database is composed of a number of LSDBs (nearly 200) on one site that has excellent graphics (as has been reported at MDI meetings [Minoshima et al. 2000]).
Updates and Hit Rates
To be useful, LSDBs must contain current data. We found 38% of databases last updated in the previous 6 mo, 10% in the first 6 mo of 2001, and 12% in 2000. A date of last update was not shown in 36% of LSDBs. This leaves the user wondering how current the information is.
Of the LSDBs examined, only 34% had counters that were publicly visible. An e-mail survey of database curators to those without visible counters elicited responses from 55% of those asked. Of these, 75% had hidden counters and provided user statistics to the authors. A counter, visible or hidden, can be a useful indicator of visitors to an LSDB and therefore an indicator of whether the site is of value to the community. Of those databases assessed, approximately half were accessed less than 100 times per month and half accessed >100 times per month. As an example, the PAH database, which is a database that we rate with a high score, is accessed an average of 750 times per month. It is difficult to gauge the exact usage of databases because it depends on how the counter is set up. For example, a counter may count each and every visit to the site, possibly many times per day from one user, or it may count each individual “user” only once per day or once per month; thus, comparison of hit rates among databases is not feasible.
Close to an Ideal LSDB?
From the compilation of all 94 websites representing 262 LSDBs currently available for examination, we tried to draw a scheme based on the “best” design, structure, and content of a database from the viewpoint of the user. Data should be organized and stored in such a way that clinicians as well as biologists can easily access them. This format and content is shown in Table 4.
A Ideal LSDB Homepage
Proposal of a Unified Network Scheme for Collecting Genotypes and Phenotypes
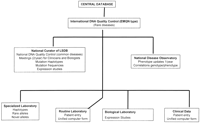
If we want to attempt to obtain complete and reliable records, including clinical and biochemical data on each patient, for each novel submitted mutation (clearly not the case in the literature or in the databases), the only way is to share a common entry form at the time of clinical diagnosis and biological experimentation. Hospitals and laboratories use a number of different computing systems to record and store clinical or biological data, none of them currently adapted to genetic disease and without any link between them. Consequently, enormous amounts of data that are never seen by potential users remain unknown and are lost to knowledge. We suggest the creation of common software that could be used by both clinicians and researchers for the same patient and would be flexible enough to be adapted to each type of genetic disease and to each type of gene by specific curators (Fig.7).
Proposal of a unified network scheme for collecting and reporting genotypes and phenotypes (adapted to each gene and disease).
This system would have to be combined with the creation of national and international networks that could ensure the quality of genotype detection and reporting and would ensure an appropriate link between phenotypes and genotypes for each patient affected with a specific genetic defect. The system would be upgraded depending on the evolution of biological and clinical data. The unified scheme would also promote the collaboration between clinicians and scientists, which will be absolutely necessary to properly handle the mass of information on the human genome that will soon accumulate.
DISCUSSION
Community health relies on the best access to current knowledge both to improve research questions and to provide maximal health care. Because variation in the genome has been said to affect 60% of individuals in a lifetime (Czeizel and Sankaranarayanan 1984), it is clear that society should spend considerable time and resources documenting this variation, particularly that affecting health. To this end, particular groups of individuals or single individuals have collected mutations in their gene of interest to help in their research or clinic and have shared them with others by publication or on the World Wide Web. These databases have been referred to as LSDBs.
Because of the importance of complete and accurate variation information, we documented in detail the content of 94 databases that represented the type of documentation currently given in such databases. It is almost impossible to discuss in detail the 80 characteristics examined; thus, only key characteristics will be discussed.
First, the most striking finding was the extreme variation between databases in all 80 characteristics examined and the number of characteristics appearing. All the characteristics are useful; thus, an ideal database should theoretically carry them all. However, the variation appearing in LSDBs can be contributed to factors such as interests of the curator, computer program literacy, funds and hands available, time of creation, and the lack of general guidelines and suitable off-the-shelf software.
Second, crucial signs for a more uniform and useful set of databases in the future look promising. The HUGO-MDI guidelines were used by 29% of databases examined in the development of their database. HUGO-MDI nomenclature was used by 42%, and the recommended allele variant entry form was used by 24% of databases that allowed submission of data (53%) as an important activity. Polymorphisms are becoming more important in the study of disease and ill health; 40% of LSDBs now contain polymorphism data and the number is increasing. These numbers are promising and likely represent in the main recently established databases, with earlier ones essentially developing in a vacuum.
Next, curation is important, and it is obvious that the 18% of databases operated by a consortium have a greater chance of survival and adequate attention being given to them. In the case in which there is more than one database per gene (22 genes), curation is clearly split, and in an environment of limited funds this is illogical and funding bodies are less likely to fund such databases.
A vital characteristic that would enhance the rate of new database creation would be the availability of “off-the-shelf” tailor-made software. Software needs to be able to allow collection, correction, and review and be able to store the data, both published and unpublished. This software needs two main functionalities: to allow operation by official curators, the software either remote or in a central facility, and to allow permanent storage of data. Such visionary software has been developed as Mutation View; however, the individual databases are operated by a central group of curators who are not necessarily expert in the gene at present. There have been recent attempts to create such software, but because of their recent creation they are not widely used outside their creator's institute; however, they do run a number of databases. For example, in February 2002 UMD software (Beroud et al. 2000) had 15 LSDBs running on this software and nine databases in development;MUTbase software (Riikonen and Vihinen 1999) runs 18 LSDBs, and there are six in development; MuStaR (Brown and McKie 2000 ) runs only two LSDBs and has two in development, even though the authors found this software the easiest to use for a novice (C. Beroud, M. Vihinen, and A. Brown, pers. comm. from software developers). Further enhancements are needed to make them more desirable to groups outside the developer's institute. Currently, some members of the HGVS are attempting to procure funds to design and build appropriate software. This funding could be from granting bodies or nondirected commercial funds to build such software.
Ease of use is clearly an important characteristic for users as well as curators. The ideal database, of course, would automatically include the search capability that was present in 35% of the databases. There are some excellent examples that we find easy to use that people can use as models for their activities: Phenylalanine Hydroxylase Knowledge Base, Blood Group Antigen Mutation Database, and A Syllabus of Human Haemoglobin Variants. For usability, these scored a 9 in our scores given out of 10 above.
Having described the content and other aspects of a representative sample of all current LSDBs, we are now in a position to accurately define what is needed for what is essentially the collective opinion of >100 curators. Clearly, the data in each of these databases are extensive and they have often been referred to as a knowledge bases, that is, being inch wide in span of genes but mile deep with information. This is in contrast to lists of mutations in central or general databases that have been described as mile wide and inch deep.
In an ideal world, all the components found in this set of databases would be ideal (Table 4), even though some curators may, as a result of lack of time or information, be unable to fulfill all fields and criteria. Such a presence of all characteristics would allow LSDBs to move toward uniformity. Second, if all fields and criteria are to be met, extensive research time needs to be available, as well as time to gather the information from the literature, patient records, and laboratory books. Ideally, this would mean the availability of a curator that could be part-time.
Software is needed to accommodate this ideal scenario and needs to be publicly available and extremely simple to use to encourage its use and have uniformity of data. The data collected needs to be stored in a permanent database and not dependent on the funding of the curator. There also needs to be some coordination, encouragement, and drive to fulfill these needs. Finally, there is a need for all this to be publicly available free of charge on the Internet.
How do we move forward to achieve this ideal of a composite and complete database ultimately of all genes (mile wide) and with extensive information (mile deep)? The first discussion of this objective began in 1994, when what later became known as the HUGO Mutation Database Initiative (now known as the Human Genome Variation Society) was convened (Cotton 2000). Numerous problems have been identified and solutions suggested in the intervening period, such as nomenclature (Antonarakis and the Nomenclature Working Group 1998, Den Dunnen and Antonarakis 2000), suggested content and quality control (Cotton and Horaitis 2000, Scriver et al. 1999, Scriver et al. 2000), and variation submission forms (allele variant entry form; see the HGVS website). From our survey here, we can see that these guidelines are beginning to be accepted and used in the design of new databases. For example, 42% of LSDBs use the HUGO-MDI nomenclature, 24% of data submissions to LSDBs are via the HUGO-MDI allele variant entry form, and 29% of LSDBs follow the complete HUGO-MDI guidelines. These are a few steps closer to the ideal database. This society aims to facilitate collection, storage, and delivery of variation information.
To further enhance mutation collection, an overall mutation collection system (HGVSYS) is being developed as a joint international effort between groups that comprise a receiving and reviewing station (Toronto; WayStation), a set of LSDB curators and others to act as reviewers or “gene editors” for data submitted, a modified HGVbase (Warehouse; Stockholm) to act as a permanent storage database, and a coordinating office (Melbourne). Incentives are being offered to induce those defining variation to submit their mutations to the system. These include a journal citation in Human Mutation, as well as a PubMed ID. This system will begin operations around mid-April 2002.
Finally, as in many areas of medical research today, there may be ethical barriers to collection of all variation; however, the existence of so many databases today indicates that it may not be a problem. Perhaps this is because the information included for each mutation in today's LSDB is similar to that provided in journal publications. If we are to move ahead and make the most of variation described in the human genome, the most essential ingredients are the ability of a committed group to work together toward the ideal and adequate funding to build and operate the system. We believe we have made steps toward this end. We hope interested individuals will use this as a guide and join us.
METHODS
We examined websites containing LSDBs of mutations of individual genes available through HGVS or MutRes websites. We, the authors, are curators of the HGVS website. This site has an extensive list of LSDBs with their URLs that are routinely updated as LSDB curators submit new databases to the list. There may be a few databases available and not included on the list that we have not been able to locate yet, but it is impossible to know for sure. The MutRes site is another extensive list curated by another HGVS member. The HGVS list may be viewed on the HGVS website. We were able to access a total of 262 genes with one LSDB for each on 94 websites. When a website contained more than one LSDB, they were found to have the same fields; thus, only one LSDB from each website was chosen for study, making a study number of 94 (i.e., equal to the number of accessible websites). LSDBs were examined for the presence or absence (yes/no) of 78 content criteria describing the structure, content, or overall ease-of-use of the database in the period from June 21 to July 18, 2001. Two extra criteria (date of last update and number of hits per month) were reviewed in December 2001, making the total number 80. These criteria have been summarized in Table 2. Complete data are available on a flat-file format atwww.genome.org (See Supplementary Table A), which is the raw data for the study.
WEB SITE REFERENCES
http://131.113.190.126/index.html; Keio database.
http://archive.uwcm.ac.uk/uwcm/mg/hgmd0.html; Human Gene Mutation Database (HGMD).
http://data.mch.mcgill.ca/pahdb_new; Phenylalanine Hydroxylase Knowledge Base.
http://globin.cse.psu.edu; Globin gene server.
http://hgvbase.cgb.ki.se; HGVbase.
http://srs.ebi.ac.uk; European Bioinformatics Institute.
http://www.bioc.aecom.yu.edu/bgmut/index.htm; Blood Group Antigen Gene Mutation database.
http://www.centralmutations.org; WayStation pilot system.
http://www.genomic.unimelb.edu.au/mdi/entry.html; HUGO MDI allele variant entry form.
http://www.genomic.unimelb.edu.au/mdi/dblist/glsdb.html; HUGO MDI lists of mutations.
http://www.gen.emory.edu/mitomap.html; MITOMAP.
http://www.hgu.mrc.ac.uk/Softdata/Mustar; MuStaR.
http://www.umd.necker.fr; Universal Mutation Database (UMD).
http://www.uta.fi/imt/bioinfo; MUTbase.
http://www.uta.fi/imt/bioinfo/IDdiagnostics; Directory of Immunodeficiency DNA Laboratories.
http://www.wiley.com/genetics/HUMUMDI; Special Issue: The HUGO Mutation Database Inititiative, vol. 15, no. 1, 2001.
http://www3.ncbi.nlm.nih.gov/omim/; Online Mendelian Inheritance in Man (OMIM).
Acknowledgments
The authors thank The University of Melbourne, the Université Montpellier, The Human Genome Organization, and the March of Dimes for financial support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵4 Corresponding author.
-
E-MAIL cotton{at}ariel.ucs.unimelb.edu.au.; Fax +61-3-9288-2989.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.217702.
-
- Received October 5, 2001.
- Accepted March 15, 2002.
- Cold Spring Harbor Laboratory Press