Functional Diversity of Mx Proteins: Variations on a Theme of Host Resistance to Infection
In vertebrates, host defense against pathogens is mediated by two general systems: innate and acquired immunity. Innate immunity constitutes the first line of defense, providing a rapid response by the expression of germ-line encoded proteins that preexist or are induced within hours of infection. Adaptive immunity is a slower, yet highly specific response mediated by B and T lymphocytes that confers effective and long-lasting protection against infection. Adaptive immunity is based on the generation of a large repertoire of antigen-recognition receptors by somatic gene rearrangement. Diversity has been considered the hallmark of adaptive immunity. In the last few years, however, evidence has accumulated supporting the importance of diversity (probably as a response to selective pressures) even within innate immunity (Hoffmann et al. 1999). Moreover, differences in innate immune mechanisms have been shown to be critical in host susceptibility to infection (Cooke and Hill 2001), making this an area of intense research.
The interferon-induced Mx1 protein is one of the best studied determinants of innate immunity to viral infection. In 1962, Lindenmann showed that the inbred mouse strain A2G is resistant to doses of mouse-adapted influenza virus that are lethal to other inbred strains (Lindenmann 1962). This was a particularly interesting observation because innate resistance in A2G mice was dependent on a single dominant locus, named Mx1, that was expressed in a variety of cell types ranging from macrophages to hepatocytes and was exquisitely specific for orthomyxoviruses. Subsequent studies showed that the specific resistance ofMx1+ murine cells to influenza viruses is attributable to the IFN-induced protein Mx1, and that after virus infection, the Mx1 protein is rapidly expressed in the nuclei of cells in the area where virus replication occurs, thus blocking viral spread (Arnheiter et al. 1996). The presence of a natural resistance gene to influenza was intriguing because mice are not natural hosts for orthomyxoviruses. Soon it became clear that Mx1 was the first member of a small gene family present in all vertebrate species from fish to men, and that the spectrum of antiviral activity was much larger than initially appreciated. However, the exact mechanism of action of Mx proteins is still a matter of debate, and it is not clear whether the antiviral activity of Mx is a luxurious accident of some undefined cellular function or has evolved to inhibit in each species a set of species-specific pathogens.
In this issue, Watanabe's group describes a series of Mxalleles in chicken breeds some of which encode proteins with antiviral activity against influenza and vesicular stomatitis virus (VSV) (Ko et al. 2002). These findings are interesting from several perspectives. First, they close the chain of observations initiated with the discovery of the innate resistance in A2G mice to a mouse-adapted influenza virus by demonstrating, as has been speculated to exist, the presence of an active Mx protein in a species that functions as a reservoir for the virus. Second, they highlight the versatility of the antiviral function of the Mx protein that is associated with a single Ser631Asn substitution present in about 50% of the breeds studied. Third, these findings also trigger important questions: Is there a physiological function for Mx proteins? Is there a common theme in the mechanism of action of Mx against viruses? What are the mechanisms that fashioned the Mx antiviral activity?
Most species have one to three Mx protein isoforms with different antiviral activities and intracellular localization. The prototype protein, mouse Mx1, as well as other rodent isoforms including rat Mx1, accumulate in the nucleus. In contrast, other isoforms as well as Mx proteins of humans and most other species are localized in the cytoplasm. Expression of Mx proteins is stimulated by IFNα/β or by viral infection irrespective of their activity. In fact, Mx proteins present a wide range of antiviral activity: nuclear Mx1 proteins inhibit principally orthomyxoviruses; in contrast, the cytoplasmic isoforms inhibit RNA viruses of the most diverse families, whereas some isoforms lack detectable antiviral function (Table 1).
Intracellular Localization and Antiviral Spectrum for Selected Mx Proteins
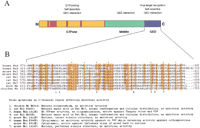
What is the structural basis of Mx function? Mx proteins belong to a superfamily of proteins, the large GTPases, which includes the dynamins, the products encoded by the Drosophila shibire, the yeast vacuolar sorting protein Vps1p, and the GTP-binding protein fromArabidopsis thaliana (van der Bliek 1999). Members of this family are present in a variety of cell locations where they perform a range of functions including endocytosis, intracellular vesicle transport, and mitochondria distribution. These proteins present modular organization characterized by at least three distinct functional domains with varied degrees of sequence conservation underlying their ability to self-assemble into higher order structures that resemble rings and helical stacks of rings, as in dynamin and Mx proteins (van der Bliek 1999). In particular, Mx proteins present a highly conserved N-terminal GTPase domain of ∼300 amino acids, a “middle” domain of ∼150 amino acids, and a GTPase effector domain (GED) of ∼100 amino acids including two leucine zippers that have the capacity to form amphipathic α-helices (Fig.1A). Studies with human MxA showed that the GED is able to specifically contact the middle domain, and that this interaction is critical to constitute a functional GTPase domain, as well as for oligomerization (Schumacher and Staeheli 1998; Di Paolo et al. 1999). Despite its functional and structural role that might be expected to restrict the amount of sequence variation among the different Mx proteins, the carboxy-terminal region shows 22% sequence identity as opposed to 38% identity in the remaining sequence (Fig.1B). This would indicate that these sequence variations underlie functional differences between the different Mx isoforms as well as with other members of the dynamin family. Indeed, divergent sequences in the carboxy-terminus on other members of the dynamin family are thought to control specific localization and functional characteristics of individual proteins, including interactions with cellular proteins (Floyd and De Camilli 1998). It is conceivable that a specialized function of Mx proteins in viral defense has evolved as a result of a direct interaction between the carboxy-terminus of individual Mx proteins and species-specific viral pathogens. Such evolutionary mechanism for host-resistance genes has been shown to prevail in plants, in which resistance to bacterial and viral pathogens is mediated by gene-for-gene interactions between corresponding genes for resistance and avirulence in host and pathogen, respectively (Erickson et al. 1999; Bergelson et al. 2001).
A: Domain structure and function of Mx proteins. Red stripes indicate the tripartite GTP-binding consensus elements, whereas the pink square indicates the ‘dynamin family signature’ consensus sequence that is present in all members of the dynamin family. GED stands for GTPase effector domain. B: Sequence alignment of the GED domain in selected Mx proteins. Red dots indicate conserved leucine residues participating in leucine-zipper domains. Sequences were aligned using the Clustal V program and the presence of seven of nine identical residues was highlighted using theBoxshade program. Essential residues for Mx activity identified in the GED domain are numbered 1 to 7.
The antiviral properties of Mx proteins differ and are influenced by the intracellular localization of the individual proteins as well as by the pathogen, underscoring their functional diversity (Table 1). In addition, several reports support a direct interaction of Mx proteins and various viral targets. Mx1 proteins accumulate in the nucleus, which is the site of orthomyxovirus transcription and replication. Here they block primary transcription of influenza virus and Thogoto virus, probably via an interaction of mouse Mx1 and the PB2 subunit of the influenza polymerase (Stranden et al. 1993). Rodent Mx2 proteins are cytoplasmic. They inhibit vesicular stomatitis virus (VSV,Rhabdoviridae) and several members of theBunyaviridae family that typically replicate in the cytoplasm, at an early stage of the viral cycle. However, when Mx2 proteins are translocated into the nucleus, they acquire activity against influenza whereas Mx1 becomes inactive if moved into the cytoplasm, indicating that the sites for VSV or influenza recognition are distinct. The cytoplasmic protein human MxA has a broader spectrum of antiviral activity. As Mx1, MxA inhibits orthomyxoviruses replication. However, in this case the block is at the posttranscriptional level through an interaction with the nucleocapsid NP protein of Thogoto virus that prevents the infecting particles from entering the cell nucleus (Kochs and Haller 1999a,b). MxA also inhibits the multiplication of measles virus; however, in this case the protective effect of MxA is cell-type specific blocking either viral RNA synthesis or synthesis of viral glycoproteins, depending on the cell line used (Schneider-Schaulies et al. 1994). Remarkably, the antiviral specificity of MxA is extended to positive strand RNA viruses, including Semliki Forest virus (Togaviridae). These pathogens are inhibited early in their replicative cycle by a block that targets viral components other than structural proteins (Landis et al. 1998).
What are the determinants of antiviral activity? GTP-binding and proper subcellular localization are two requisites, but they are not sufficient. Extensive mutation analysis and, now functional analysis of Mx variants emphasize the critical role of the GED domain in GTPase activity, polymer formation, and antiviral activity (Fig. 1B). Interestingly, introduction of a L612K substitution in human MxA abolishes both GTPase activity and the ability to form polymers while retaining its antiviral activity against Thogoto virus and VSV. This would indicate that high-molecular weight MxA oligomers represent a storage form, whereas GTP-bound MxA monomers are the active form of MxA (Janzen et al. 2000). In contrast, a number of single point mutations, summarized in Figure 1B, result in loss of antiviral activity despite maintaining adequate subcellular localization and the predicted overall helical structure of the carboxy domain. A noteworthy MxA mutant is the human E645R protein that has lost activity against VSV while retaining activity against influenza and Thogoto virus, further supporting the notion of a direct interaction of the protein with different viruses via the carboxy-terminus. Also remarkable is the fact that the active (Mx2) and inactive (Mx3) isoforms of the murine Mx protein differ by eight amino acids, however, antiviral activity is only abrogated by amino acid changes occurring in the GED domain. Moreover, Ko et al. (2002) report that among a number of naturally occurring mutations in chicken Mx only the N631S mutation in the GED domain resulted in loss of antiviral function providing further evidence of the key role of the GED domain in antiviral activity. These findings, together with the observation that GTP-bound MxA cosediments with viral nucleocapsids, are consistent with a model in which the antiviral activity of Mx proteins depends on specific interactions between the GED and specific viral structures (Janzen et al. 2000; Kochs et al. 2002). These varied interactions do not preclude, however, a common antiviral mechanism of action. Some exciting insights into a possible mechanism of action have been provided from the recent discovery that Mx1 protein is associated with components of the SUMO-1 system of promyelocytic leukemia protein nuclear bodies (PML NBs) (Engelhardt et al. 2001). It is conceivable that subsequent to binding, Mx proteins bring viral targets to PML NBs known to play an important role in proteasome-mediated degradation of ubiquitinated proteins and to be involved in the cell defense against varied viral infections (Everett et al. 1997; Anton et al. 1999).
The presence of inactive variants of host-resistance genes is always a matter of debate. In wild mice that possess two Mx genes, the frequency of an inactive Mx1 allele is 50% (Haller et al. 1987), as it is for chicken Mx in the 18 breeds investigated by Ko et al. (2002). In terms of evolution, it has been proposed that increased resistance to infection would be costly in terms of other traits otherwise, the frequency of susceptibility alleles might be expected to decrease and resistance would be monomorphic in the presence of disease (Boots and Bowers 1999). Possible detrimental effects of Mx gene expression in specific cell populations was suspected in the production of transgenic mice in which high levels of Mx1 or MxA proteins were shown to confer resistance to viral infections (Kolb et al. 1992; Pavlovic et al. 1995). It was clear, however, that expression of the proteins was not tolerated in some cell populations including hepatocytes, probably due to inadequate expression of the transgene in these cells or embryonic lethality. More recently, it was reported that MxA is overexpressed in lymphoblastoid cell lines from patients with Fanconi anemia (FA) (Li and Youssoufian 1997). FA is a group of genetic disorders characterized by congenital defects, bone marrow failure, cancer susceptibility, and abnormal genomic instability in cells. All FA subtypes seem to converge to a common pathway characterized by overexpression of MxA that is sufficient to induce apoptosis, indicating that loose control of Mx gene expression is deleterious for the organism. Alternative possibilities for the presence of Mx inactive variants would be the acquisition of advantages against different pathogens not yet characterized. In that respect, it would be interesting to retrieve the geographical location of the susceptible lines, as well as the prevalence of pathogens in the respective areas, to test the hypothesis of new antiviral activities. Again, in plant systems, allelic variability has been proposed to underlie the adaptive acquisition of novel resistance alleles, and correlations between the geographical distribution of resistance allele with specific pathogens have been shown (Bergelson et al. 2001). Other models accounting for the maintenance of susceptibility alleles in a population are described as the “hitch-hiking effect”, in which the presence of a favorable gene conferring a heterozygous advantage maintains tightly linked susceptibility genes in the population (Smith and Haigh 1974; Malo et al. 1994). However, the possibility that Mx proteins fulfill a cellular function involved in cellular trafficking and/or in stress responses (Horisberger 1992) apart from their documented antiviral effects remains valid.
Currently, major efforts are underway to identify and catalog single-nucleotide polymorphism in humans and economically important species. Mining for variations in Mx genes together with systematic screening of their antiviral properties in in vitro systems seems a promising avenue to further our understanding of the functional diversity found within Mx proteins. This will provide a rich opportunity for the study of the evolution of host-virus interactions and a unique chance to study both evolutionary and genetic aspects of the host antiviral functions.
AKNOWLEDGMENTS
We are grateful to Dennis Bulman and Douglas Gray for critical reading of the manuscript and Danielle Malo for stimulating discussions throughout the preparation of this work.
Footnotes
-
↵2 Corresponding author.
-
E-MAIL svidal{at}uottawa.ca; FAX (613) 562-5452.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.20102.
- Cold Spring Harbor Laboratory Press