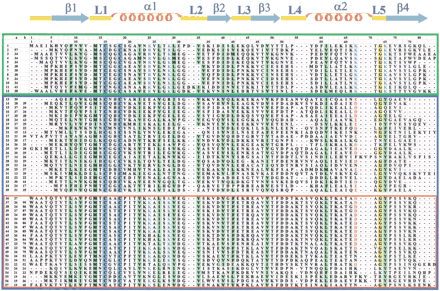
Sequence alignment of the Atx1 amino acid sequence from yeast S. cerevisiae with the metallochaperone homologs located in the present research. At the top, amino acid numbering is reported including gaps (first line) and according to the sequence of Atx1 (second line). The two metal-binding cysteines are shaded in blue. Positions where hydrophobic residues are conserved are shaded in green. Key positive (Arg and Lys) and negative (Glu and Asp) residues are indicated in blue and in red, respectively. The boxes include eukaryotic metallochaperone sequences (green), and bacterial, CopZ-like and MerP-like, sequences (blue). The red box includes the subgroup of MerP-like sequences. The secondary structure elements reported above the alignment are referred to Atx1. In column a we report sequence identity to Atx1 (sequence 1); in column b we report identity to sequence 13 and sequence 36 for CopZ-like and MerP-like sequences, respectively. Eukaryotic metallochaperones: 1 gi‖6730164 Saccharomyces cerevisiae(Atx1); 2 gi‖7492713 Schizosaccharomyces pombe;3 gi‖11290108 Oryza sativa; 4 gi‖11290106 Glycine max; 5 gi‖15228869 Arabidopsis thaliana;6 gi‖1945365 Homo sapiens; 7 gi‖6013208Canis familiaris; 8 gi‖7531050 Ovis aries;9 gi‖7531046 Mus musculus; 10 gi‖7531044Rattus norvegicus; 11 gi‖4165309 Caenorhabditis elegans; 12 gi‖7296474 Drosophila melanogaster;Bacterial and archaeal soluble metal transporters: CopZ-like: 13 gi‖5107576 Enterococcus hirae (CopZ); 14gi‖7429061 Bacillus subtilis (CopZ); 15 gi‖1477773Helicobacter pylori, strain A68; 16 gi‖9789743H. pylori, strain J99; 17 gi‖3121871Helicobacter felis; 18 gi‖7471711 Deinococcus radiodurans; 19 gi‖7462405 Thermotoga maritima;20 gi‖11346720 Campylobacter jejuni; 21gi‖13357760 Ureaplasma urealyticum; 22 gi‖8894834Streptomyces coelicolor; 23 gi‖9965436Streptococcus mutans; 24 gi‖13622769Streptococcus pyogenes; 25 gi‖11353981Neisseria meningitidis, serogroup A strain Z2491; 26gi‖11353792 N. meningitidis, serogroup B strain MC58;27 gi‖14600385 Aeropyrum pernix; 28gi‖14195314 Haemophilus influenzae; 29 gi‖12722322Pasteurella multocida; 30 gi‖12723757Lactococcus lactis; 31 gi‖12725117 L. lactis II; 32 gi‖10580283 Halobacterium sp.; 33gi‖10173169 Bacillus halodurans; 34 gi‖11349934Pseudomonas aeruginosa; 35 gi‖8388756Pseudomonas syringae; MerP-like: 36 gi‖127010Shigella flexneri (MerP); 37 gi‖2944141Pseudomonas stutzeri I; 38 gi‖2947088 P. stutzeri II; 39 gi‖2498542 Alcaligenes sp.;40 gi‖127008 P. aeruginosa, plasmid pVS1; 41gi‖2498544 Pseudomonas fluorescens; 42 gi‖4572444Sphingomonas paucimobilis; 43 gi‖127009Serratia marcescens; 44 gi‖4572382 E. coli;45 gi‖2498541 Acinetobacter calcoaceticus;46 gi‖2498543 Enterobacter cloacae; 47gi‖2052180 Pseudomonas sp., strain KHP41; 48gi‖2159997 Pseudomonas sp., strain K-62; 49gi‖6689527 Xanthomonas campestris; 50 gi‖2498545Shewanella putrefaciens; 51 gi‖2935549Pseudomonas alcaligenes; 52 gi‖2765117Thiobacillus sp.; 53 gi‖14195504 H. influenzae; 54 gi‖12721567 P. multocida;55 gi‖10640685 Thermoplasma acidophilum; 56gi‖14324466 Thermoplasma volcanium; 57 gi‖11496143Pseudoalteromonas haloplanktis.











