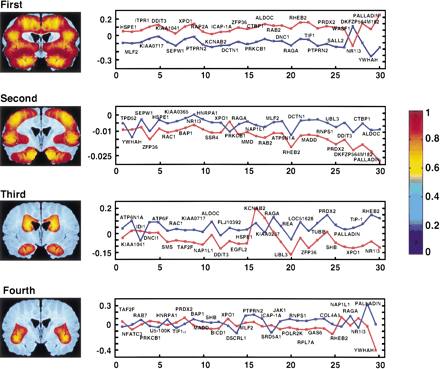
SVD delineates anatomical regions of the brain. The conjoint matrix resulting from the top 120 genes most strongly (P ∼ 0.05) differentially expressed between the normal and Alzheimer's hemisections was analyzed. The spatial patterns resulting from the first, second, third, and fourth PCs are shown. Alongside are the first 30 members of the corresponding gene vectors. The ordinate represents the contribution by the relevant gene to the variation of the vector spatial pattern, whereas the abscissa represents the genes in decreasing order of significance of differential expression. The genes are indicated by UniGene symbol or name. Normal: red, Alzheimer's: blue. The first component is uniformly expressed over the brain, and represents an image of average gene expression differences between the samples. The second component is largely restricted to cortex, the third to both the tail of the caudate and the hippocampus, and the fourth to the insular cortex. The level of expression of the relevant gene vector in the spatial patterns can be deduced by reference to the pseudocolor scale (right). Imaging software smoothed the expression patterns over the voxels, and the hemisection was reflected along the midline for the figure, giving bilateral symmetry.











