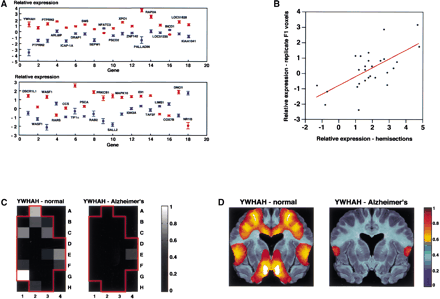
Genes whose average expression across voxels is significantly different in the normal compared to the Alzheimer's brain. (A) Graph showing mean expression levels across the 24 voxels in the normal and Alzheimer's hemisections on a logarithmic scale (log2) (±SEM). Normal: red, Alzheimer's: blue. The genes are ranked from most (gene 1) to least significant (gene 36, P< 10−7). Two genes of the differentially expressed subset, PTPRN2 (genes 2 and 3, upper row) and WASF1 (genes 2 and 3,lower row), were present as duplicate spots on the microarrays, and give an independent assessment of within array replicability. (B) Scatterplot comparing the mean expression differences between the normal and Alzheimer's disease brains based on the hemisection data and the replicate F1 voxel data. Expression differences are shown using the logarithm (log2) of the gene expression ratios between the normal and diseased specimens. The genes employed in the scatterplot are those judged significantly (P< 10−7) different when averaged across the whole hemisections and which are also present on the 5000 gene microarray used to analyze the replicate F1 voxels. A total of 27 genes resulted (YWHAH, PTPRN2, ARL6IP, ICAP-1A, DRAP1, SMS, SEPW1, NFATC3, PSCD2, XPO1, ZNF142, PALLADIN, RAP2A, BICD1, LOC51628, DSCR1L1, WASF1, RARS, CCS, TIF1α, PRKCB1, SALL2, MAPK10, IDH3A, IDI1, TAF2F, DNCI1). There was a highly significant correlation between the data from the hemisections and the F1 voxels (r = 0.65, F[1,25] = 18.34, P = 0.0002). The best fit using least squares linear regression is shown. (C) An example of the spatial expression pattern of a gene (YWHAH) whose expression is significantly greater in the normal compared to the Alzheimer's brain. The level of gene expression can be deduced by reference to the scale on the right. (D) YWHAH expression patterns after smoothing over voxels using imaging software, and projecting onto the relevant neuroanatomy. The resulting images were reflected along the midline for the figure, giving bilateral symmetry.











