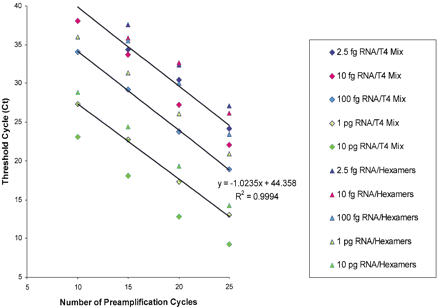
Random hexamers versus T4 Mix priming with Sensiscript. Twenty narograms, 2 ng, 200 pg, 20 pg, and 5 pg of total tracheal RNA (i.e., RNA equivalent to ∼2000, 200, 20, 2, and 0.5 cells) were used in cDNA synthesis with Sensiscript as described in Methods using either random hexamers or T4 Mix with 200 gene-specific primer sets. Aliquots of RT reactions were PCR-amplified with the T4 Mix for 10, 15, 20, and 25 cycles as described in Methods, and 0.05% of each reaction used in real-time PCR to quantify the levels of GAPDH. The number of preamplification cycles in the multiplex step was consistently and inversely related to cycle threshold for GAPDH in the real-time PCR step. Both gene-specific and random hexamers efficiently primed the first strand cDNA with Sensiscript, although gene-specific priming was much more efficient and resulted in consistently lower Cts for GAPDH. Multiplex preamplification with T4 Mix was also efficient and linear and produced slopes of −1.0 (at least for GAPDH).











