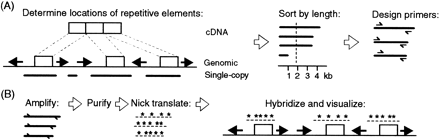
The process for developing and producing scFISH probes. (A) Computational sequence analysis of genomic sequence. Genomic sequence is compared with a database of repetitive sequences to identify the positions of repeat elements (left arrows or right arrows), thus delineating the boundaries of single-copy intervals (solid lines). These deduced sequences are then compared with the genome sequence to detect potential gene family members, pseudogenes, and other complex low-copy repeats (data not shown). The intervals are then sorted according to their respective lengths, and primers (⇀ and ↽) are optimized to amplify intervals with lengths that exceed a threshold (i.e., 2 kb). (B) Single-copy intervals are amplified by long PCR of high molecular weight genomic DNA. The PCR products are then purified, labeled by nick translation (underlined asterisks), hybridized, detected with a fluorescent antibody that recognizes the labeled nucleotide, and visualized by fluorescence microscopy.











