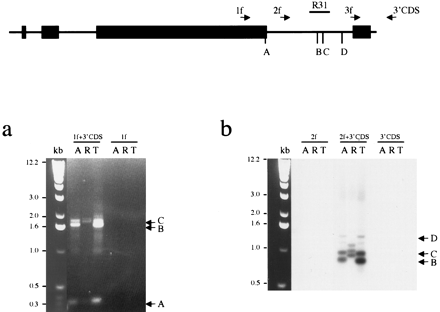
Mapping the vole Xist 3′ end. The schematic represents theXist exons 5–8. The location of primers used for 3′ RACE is indicated with arrows. The positions of major polyadenylated 3′ ends ofXist transcripts (A, B, C, D) and probe R31 used for hybridization are shown. (a) 3′ RACE amplification of M. arvalis (A) and M. kirgisorum (T) total RNA, and M. rossiaemeridionalis (R) poly A+ RNA with combination of gene-specific and universal primers, 1f +3′CDS, 2f +3′CDS, 3f +3′CDS. 1f, 2f and 3′CDS primers only were used as negative control to assure specificity of amplification. Ethidium bromide stained gel is shown for primer pair 1f +3′CDS and 1f control. Three major bands (A, B, C) are indicated with arrows. (b) Southern blot hybridization of Xist probe R31 is shown for primer pair 2f +3′CDS and controls to prove specificity of amplified fragments. Bands corresponding to the 3′ ends of Xist (B, C, D) are indicated with arrows.











