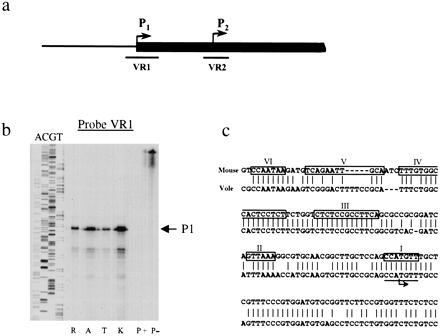
Mapping the vole Xist transcription start site. (a) A scheme of vole Xist, including the putative transcription start sites and the positions of the probes used in nuclease protection assay. (b) Nuclease protection assay was used to detect the position of transcription start site. Antisense riboprobes were synthesized spanning the predicted P1 and P2 start sites. 10 μg of total RNA from fibroblast cell cultures was hybridized to radiolabeled probe and digested with mung bean nuclease. Probe-, undigested probe; probe +, digested probe after hybridization to yeast RNA. Lanes R,A,T,K are forM. rossiaemeridionalis, M. arvalis, M. kirgisorum, and M. transcaspicus RNA, respectively. (P1) The band corresponding to the size of P1 start site. A sequencing ladder of known fragment is shown alongside to estimate the position of start site. The data for P1 riboprobe only is shown. (c) Sequence comparison of Xist minimal promoter between vole and mouse. Consensus initiator sequence is underlined, and the position of transcriptional start site is indicated by arrow. Conserved promoter elements I–VI are boxed (Sheardown et al. 1997b).











