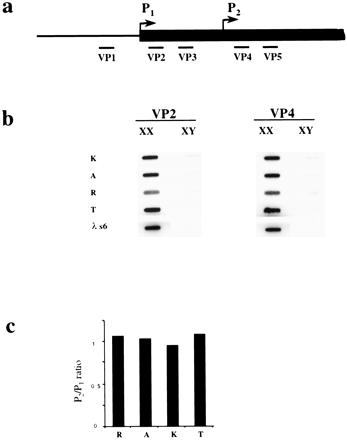
Xist transcription analysis. (a) A scheme of voleXist, including the putative transcription start sites and the positions of the probes used in slot blot analysis. (b) Hybridization of probes to slot blots containing 10 μg of total RNA from XX and XY kidneys. No hybridization was detected for upstream VP1 probe (data not shown). The results obtained for VP2 and VP3 and for VP4 and VP5 probes were similar, hence only representative slot blots for VP2 and VP4 probes are shown. The signal was normalized to 28S RNA probe and to lambda Xist DNA for the efficiency of hybridization. (c) Quantification of slot blot data showing the usage of vole P1 and P2 start sites. Black bars represent P2/P1 ratio for M. rossiaemeridionalis (R), M.arvalis (A),M. kirgisorum (K), and M. transcaspicus (T) RNA signal. The value close to 1 suggests that all transcripts initiate upstream the VP4 and VP5 probes.











