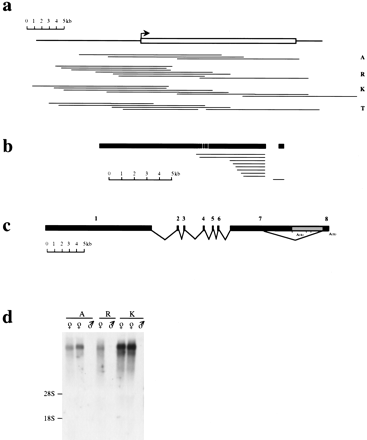
Cloning of vole Xist gene. (a) Series of λ clones isolated from genomic DNA libraries for common vole species, M. arvalis (A), M. rossiaemeridionalis (R), M. kirgisorum (K), and M. transcaspicus (T). The clone contigs have covered the whole Xist gene sequence, including extra 5′ and 3′ sequences. 5′ and 3′ boundaries of vole Xist gene were determined on the base of homology analysis with mouse. The predicted 5′ end of the gene is indicated (arrow). (b) A series of cDNA clones pulled out of M. arvalis oligo(dT) library using exon 7 and 8 probes. The size and position of clones relatively to the Xist transcript is indicated. A single clone obtained with exon 8 probe contains 0.6 kb of intron 7 sequence. Exons are shown in black with the position of introns shown in white. Exon 8 is shown distantly to the rest of the transcript to indicate the size of the intronic sequence in exon 8-contaning clone. (c) Exon-intron structure of vole Xist gene based on RT-PCR and cDNA cloning analyses. Vole Xist consists of eight exons depicted as black blocks; grey box represents long Xisttranscripts found in 3′ RACE. A (n) denotes the 3′ ends of RACE products, although classical polyadenylation sites are absent in the sequence. (d) Northern blot of M. arvalis(A), M. rossiaemeridionalis (R), and M. kirgisorum (K) total RNA hybridized to exon 1 (Rx8Pst2) probe. Expression of vole somatic Xist is female-specific in all species studied.











