A Method For Parallel, Automated, Thermal Cycling of Submicroliter Samples
Abstract
A large fraction of the cost of DNA sequencing and other DNA-analysis processes results from the reagent costs incurred during cycle sequencing or PCR. In particular, the high cost of the enzymes and dyes used in these processes often results in thermal cycling costs exceeding $0.50 per sample. In the case of high-throughput DNA sequencing, this is a significant and unnecessary expense. Improved detection efficiency of new sequencing instrumentation allows the reaction volumes for cycle sequencing to be scaled down to one-tenth of presently used volumes, resulting in at least a 10-fold decrease in the cost of this process. However, commercially available thermal cyclers and automated reaction setup devices have inherent design limitations which make handling volumes of <1 μL extremely difficult. In this paper, we describe a method for thermal cycling aimed at reliable, automated cycling of submicroliter reaction volumes.
Rapid thermal cycling of small samples containing thermally stable polymerase is required for performing PCR and for Sanger sequencing of DNA. Despite the typically small sample volumes, the cost of performing these processes on a large number of samples can be exorbitant because of the high cost of the enzyme, primers, and dyes. For example, typical cycle sequencing containing TaqFS (PE Applied Biosystems) or Thermosequenase (Amersham) enzymes in quantities and concentrations recommended by the supplier can cost in excess of $2.00 per reaction. Although most sequencing centers dilute these reagents, the costs using commercially available thermal cycling equipment cannot be lowered to less than $0.50 per sample because of minimum-volume limitations of the available instruments. This is a particularly serious problem for laboratories engaged in high-throughput DNA sequencing. Because some labs now reach throughputs in excess of 100,000 samples per day, the cost of performing the cycle sequencing reaction alone can exceed $50,000 per day. According to cost figures from one high-throughput sequencing center (Stanford DNA Sequencing and Technology Center), the sequencing reaction costs are responsible for >30% of the total cost of obtaining sequence data from a specific colony or plaque.
To reduce this potentially prohibitive cost, many centers have reduced the reaction volumes and diluted the stock enzyme/dye mix (referred to as “brew”). This is particularly appropriate now that automated capillary sequencers with high sensitivity are available (e.g., ABI 3700, PE Biosystems; MegaBACE 1000, Amersham Molecular Dynamics). Standard, full-volume reactions produce hundreds of times more products than are actually required to achieve a reasonable signal on these instruments (Meldrum et al. 2000).
Unfortunately, there are technical problems limiting both volume reduction and dilution. Evaporation of the sample during cycling and difficulties in small-volume sample handling during reaction set up have, to date, limited minimum reaction volumes to between 3 and 5 μL (12–20 μL total volume for a dye primer reaction). In addition, substantial dilution of brews can lead to degradation of sequence quality, presumably because of decreased nucleotide and enzyme concentrations in comparison to contaminant concentrations present in the template (Salas-Solano et al. 1998). Therefore, cost reduction factors of 10 or more cannot be achieved solely through brew dilution, but must be addressed by solving the small-volume limitations of this process.
A further cost benefit from reaction-volume reduction is realized for some reaction product purification methods. For example, the cost of magnetic bead-based purification is dependent on the volume of the reaction to be purified. Decreasing the reaction volume by a factor of five or 10 would presumably decrease some purification costs by the same factors.
Consequently, it is desirable to create an instrument capable of setting up and thermally cycling, without evaporation, reactions with volumes <1 μL. If this can be achieved, cost reduction and matching of product quantity to the capillary sequencer's decreased requirements can be achieved by volume reduction without the need for brew dilution. Furthermore, it would be desirable to create such an instrument with the capability for highly parallel operation and short cycling times. This would help to meet the 100,000+ samples/day throughput requirement that this process now faces, and would allow more efficient execution of protocols tailored to capillary sequencing.
Although instruments exist that are capable of very rapid thermal ramping (e.g., Perkin Elmer 9700, MJ Research) (Wittwer et al. 1989) and highly parallel operation, these devices are incapable of performing the pipetting required for initial reaction setup. In addition, these instruments typically use standard 96-well or 384-well microtiter plate formats for the reaction samples, leading to minimum-volume limitations from evaporation and liquid handling problems.
This paper describes a technique for setting up and thermally cycling submicroliter samples in a highly parallel format without evaporative losses and with reliable pipetting. The method described is designed to be automated in a highly parallel format with the goal of eventually constructing a single instrument capable of a 10,000-samples/day unattended operation.
Capillary Thermal Cycling in Disposable Tips
The use of capillaries as sample containers has long been recognized as a superior way of performing thermal cycling on small-volume samples (Wittwer et al. 1989; Meldrum 1997; Meldrum et al. 2000). The challenges posed by this technique arise in interfacing the capillary format to standard microtiter plate formats, and in accurately aspirating and dispensing reactions into and out of the capillary tubing (typically a small diameter glass tube). Commercially available capillary thermal cyclers (e.g., from Idaho Technology or Corbett Research) require that samples be transferred from a 96-well format into individual capillary tubes that are arrayed in a special format on the instruments. After cycling, the capillaries must again be individually removed from the cycler and the sample transferred back into a 96-well or 384-well format. This is a process that is not well suited to high-throughput automation in which it is essential to handle 96 or 384 capillaries at one time and with a single tool.
Although the method presented here shares similarities with these capillary thermal cycling approaches, the difference lies in the pipetting system and choice of capillaries. In this method, disposable 30-μL pipette tips are used as capillary containers to reduce disposables costs and to ease the task of automating the process. This choice of capillaries has the principal advantage of allowing the pipetting step and cycling step to occur within the same capillary: Small samples need not be transferred to a sample plate nor to a separate capillary for cycling, as is required with standard thermal cycling instrumentation. In addition, disposable tips are easily handled robotically; they can be easily heat sealed to prevent evaporation during thermal cycling, and then subsequently cut. The tapered inner diameter of the tips makes them suitable for reaction volumes ranging from 300 nL to 5 μL. Finally, 96 pipette tips can be handled in parallel in a standard 96-well format, greatly facilitating interfacing of the automation to standard input plate and product plate formats.
Multiplexing of dye primer samples might also be possible in these tips by separating the four different reactions by air slugs during aspiration, cycling the reactions thus separated in one tip, and pooling the reactions during the dispensing phase.
The process is simple: A robotic pipettor picks up 96 tips, loads and mixes template and brew, aspirates an air slug (and repeats loading if multiplexing is to be performed), melts and seals the end of the pipette tip to prevent evaporation, places the tip in a specially designed thermal cycling block, disengages from the tip, and seals the top of the tip with a plunger-style lid that forms a seal inside the tip. Cycling can then be performed while the pipettor attends to setting up other reactions. Once cycling is complete, the pipettor removes the tips from the cycler, cuts off the sealed pipette tip ends, and dispenses the products into a microtiter plate prefilled with a small volume of ethanol or water.
It should be emphasized that in the above process, the small samples are only present inside the pipette tips and are never transferred except in and out of bulk reagent reservoirs. The process, although straightforward, contains several steps critical to its success: accurate pipetting, sample mixing within the pipette tip, sealing to prevent evaporation, and rapid thermal cycling. The challenges inherent in these processes are discussed individually.
Pipetting
A major requirement for setting up small-volume sequencing reactions is the capability for liquid transfer with better than 30-nL repeatability. Although some commercial pipettors can achieve this, the thermal cycling scheme described in this paper requires the additional constraint of maintaining 30-nL repeatability with multiple fluid/air interfaces present in the pipettor tip.
All pipettors that use air as a pneumatic fluid to transfer the pipetting force from the plunger to the sample suffer from some loss of repeatability as multiple fluid slugs are aspirated. This results from an increase in the apparent viscosity of the fluid/air mixture, presumably because of surface tension effects, and interactions of the fluid/air interfaces with the capillary walls. Experiments indicate that flow velocity (at fixed pressure differential) of the aspirated fluid slugs decreases with an increasing number of interfaces. Initial tests of the pipettor system described below showed that, for a single water slug, a 1-μL plunger motion was followed almost instantaneously (<1 sec) by a 1-μL volume displacement of the fluid slug. With 10 fluid slugs present in the same tip, the 1-μL plunger displacement was also followed by a 1-μL displacement of the 10 slugs, although in this case, the time required for the slug flow was >30 sec. Severe pipetting inaccuracies can occur in the latter case because the pressure differential responsible for pipetting motion is held for a long time and thus is susceptible to minute leaks in the pipettor seals. In addition, surface tension effects at the end of the pipette tips can cause pipetting inaccuracy by holding off some of the pressure differential created by the plunger motion.
These effects can be minimized by decreasing the air volume through which the pneumatic plunger action is transmitted. In the case of a fluid interface that withstands a fractional pressure drop of ΔP/P, the volumetric pipetting error will be ΔV = V0(ΔP/P) where V0 is the air volume confined by the pipettor plunger on one side and the fluid slug on the other.
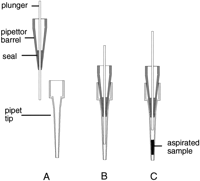
To minimize this air volume, the pipettor described in this paper is modified in such a way that the plunger exits the pipettor barrel and displaces air within the disposable pipette tip before aspiration (Fig.1). This allows the dead-air volume (the volume through which the pneumatic pipetting force is transferred) to be reduced from a minimum of 28 μL (plus any volume inside the pipettor for a regular pipettor) to 16 μL (including the pipettor volume). This reduction is particularly valuable when aspirating multiple slugs as (in addition to the increased effective viscosity described above) additional air volume is aspirated in the form of air gaps between slugs. For five slugs, this leads to 2.5–5 μL (depending on the desired size of the air gaps) of additional dead-air volume.
Diagram of pipettor configuration. The pipettor plunger slides against a seal at the bottom of the pipettor barrel and can protrude beyond the end of the barrel to displace air volume inside the disposable tips and improve pipetting accuracy.
Pipette tips are mated to the pipettor barrel by a press fit as used in handheld pipettors. Approximately 1–2 lbs of force are required for a good press fit for each tip.
Sample Mixing
If the sequencing reactions are to be set up inside the pipette tips, enzyme brew and template must be mixed adequately during pipetting. Because of the small volumes involved, mixing by expelling the samples into a plate and then reaspirating into the pipettor is not an option. Unfortunately, the low pipetting speeds (<10 mm/sec) and small tubing diameter (<1 mm) lead to a very low Reynold's number flow (R = ρVmaxd/μ ≈ 10), ensuring that the flow of the sample during pipetting will be laminar and prevent turbulent mixing from occurring. This leaves only diffusion as a reasonable mixing mechanism, which might prove to be the limiting factor for the maximum reaction volume produced with this method. Fortunately, the goal of thermal cycling is to cycle small volumes for which diffusive mixing is adequate. Furthermore, the pipetting action allows us to move the sample component to be mixed into the radial center of the fluid slug. This requires that diffusion only take place through the radial extent of the sample, typically <500 μm in a small pipette tip. The time required for diffusion of DNA and enzyme across this distance is ∼2–3 min at room temperature and will therefore occur within the first thermal cycle. Based on the performance described below, it does not appear that this mixing time has a measurable negative effect on sequencing reactions.
Sealing and Evaporation
Evaporative loss of sample during thermal cycling can occur in one of three ways. First, some amount of sample will be converted from liquid to gas phase to provide the required equilibrium vapor pressure in the surrounding air volume. At 100° C, this corresponds to 50 nL for every 10 μL of air volume. Second, if the sample container is not properly sealed, the vapor will escape and lead to continued evaporation of the sample, which usually leads to complete sample loss. Third, if one surface of the sample container is at a lower temperature than the sample, evaporative liquid transfer will occur from the sample to the cold surface. Liquid will evaporate from the sample and condense on the colder surface, changing the concentration of the sample. Although the liquid is not lost in this case, it might condense in another location from which it is irretrievable.
Capillary thermal cycling in pipette tips is particularly well suited to solving the first two of these difficulties. Polypropylene pipette tips can be effectively sealed by melting one end and by press-fitting plungers to seal against their inner diameter. Samples can be recovered by cutting the melted end, a process that is easily automated. In addition, the air volumes on either side of the sample can be made arbitrarily small, reducing evaporative loss into the enclosed air volume. The third issue, condensation, remains a problem for capillary geometry, particularly when the capillaries are sealed by unheated steel plungers, as proposed in this paper. To prevent evaporative transfer of water from the sample to the colder plunger, a substantial (2 μL) slug of water is loaded in the pipette tip just above the sample. This slug is situated in the thermal gradient at the top of the tip, and partially evaporates over the length of the cycling run. In doing this, it acts as an evaporation barrier for the sample and helps to contribute some of the equilibrium water vapor pressure to the air space above the sample, thus further limiting sample loss by evaporation.
Thermal Cycling
The long dimension of the pipette tip geometry in the direction perpendicular to the plane of the 96-tip array makes thermoelectric thermal cycling devices inappropriate for cycling pipette tips arrayed in this format. Although it is possible to build a thermoelectric cycler to cycle tips in this configuration, thermal ramp times become unacceptably long because of the time required for heat to flow in the axial direction of the pipette tip. This is particularly important if the instrument must be capable of cycling standard (5–10 μL) volumes as well as submicroliter volumes.
Conversely, the large surface-area-to-volume ratio of the tip geometry makes it ideal for cycling by means of a flowing fluid such as air or water. Water was chosen for this instrument because of its inherently higher thermal stability and uniformity resulting from its high specific heat. To achieve fast thermal ramps, three separate water baths are maintained at the temperatures required during the thermal cycling. Computer-controlled valves and pumps determine which bath is selected as the source of water to flow through the container housing the tips. For multiple arrays of tips, a single set of three baths could be made to serve several tip containers through manifolds of computer-controlled valves. Thermocouples in the tip containers can be used to determine when a ramp is complete and when the valves should be switched. The simple eight-channel prototype used for the first proof-of-concept is shown in Figure 2 to illustrate this cycling method.
Eight-channel prototype of thermal cycling bath. Circulating water enters the sealed bath at the bottom, flows around the pipette tips, and exits at the top. One of the tips is shown loaded with a sacrificial water slug (not easily visible in the figure) followed by four 500 nL slugs of loading dye for purposes of this illustration. The tips are heat sealed at the bottom end and sealed with a plunger at the top end. After cycling, the heat-sealed end is cut to remove the sample.
RESULTS
A 96-channel prototype with a custom-built automated pipettor was constructed and used for initial testing of this thermal cycling concept. This device is described in the Methods section. Although an earlier eight-channel prototype shown in Figure 2 was constructed, the sequence quality, pipetting, and thermal ramp rate results described below are from the 96-channel device (Fig.3).
96-channel automated pipettor and workspace. The pipettor is attached to an XYZ gantry, which gives it the capability to pick up disposable tips, load them with reagents, and insert them into the 96-channel cycling block, all of which are located on the workspace area.
Evaporation
Long-term evaporation effects on samples contained within sealed pipettor tips were evaluated by loading tips with 2 μL of water, a 2-μL air gap, 1 μL of sample (enzyme brew), and a further 2 μL of air. The tips were heat sealed at the sample end, sealed with a plunger at the large end, and immersed for 30 min in 100° C water. The part of the tip containing the sample was immersed in the water, while the upper part of the tip containing the plunger was out of the water and exposed to air at room temperature. After 30 min, the tips were inspected to determine what fraction of the sample remained as a liquid.
The measured volume in the sample slug after cooling was 0.99 μL, on average. The standard deviation of 0.032 μL over the six tips tested is more likely a result of measurement error than of evaporation.
Pipetting Reproducibility
The pipetting reproducibility of the tool head was analyzed using two tests. In the first test, a new set of 96 tips was fixed to the tool head. The following loading sequence was then aspirated into the pipette tips: 1000 nL loading dye (0.25% bromophenol blue, 0.25% xylene cyanol FF, 15% Ficoll [Type 400] in water), 1000 nL air, 300 nL loading dye, 500 nL air. The pipette tips were heat sealed using the tip melter. Each pipette tip was removed manually from the tool head, and the length and position of the lower slug of loading dye relative to the bottom of the pipette tip was measured using digital calipers with an accuracy of 0.0005”.
The second test attempted to simulate actual reagents in the pipette tips. Pipette tips were mounted manually onto the tool head, and were precoated with BSA (bovine serum albumin) at 1x concentration for 10 washes, and were aspirated with the following sequence: 2000 nL water, 1000 nL air, 300 nL template/brew premix, 500 nL air. The length and position of the lower slug of template/brew premix were measured relative to the bottom of the pipette tip using the digital calipers.
The measured variation (standard deviation) of the 300 nL loading dye sample volumes is 11.1% of the sample mean. Total measurement error is approximately 10%, which might account for part or all of the measured variation.
The increased viscosity in the enzyme brew samples resulted in a slightly greater variability at 15.6% of the sample mean.
Thermal Cycling Parameters
The temperature uniformity of the 96-channel thermal block was tested at a steady-state temperature of 70° C using two thermocouple wires placed inside prepared pipette tips. One of these wires was moved to sample all 96 pipette tip positions. In addition, thermal ramp times were measured (Table 1) for all relevant temperature transitions for a dye terminator protocol. Ramp time is defined as the time required for the temperature to come within 1.0° C of the final target temperature. The total cycle time spent ramping is 25 sec.
Thermal Ramp Rates
The steady-state measurements of temperature at each position showed that the maximum sample-to-sample variation in the measured temperature was 0.2° C, on the order of the rated precision of the thermocouple wire used.
Sequence Quality
Initial tests on the proposed cycling method were performed by attempting dye/terminator reactions in increasingly smaller volumes. A total of 25, 96-well plates of Arabidopsis thaliana templates were cycled using Perkin Elmer BigDye Terminator kits. Initial testing was performed using a reaction volume of 2 μL and undiluted brews. Subsequent tests were performed with smaller reactions, first with 1-μL total volume and then with 500 nL total volume. These small reactions were mixed with 5–10 μL of deionized water during expulsion from the pipette tips after cycling and purified using Sephadex G-50 (DNA Grade F, Pharmacia). The samples were then run (without any pooling) on an ABI 3700 sequencer. Initial results from these tests yielded an average of 585 Phred 20 (Ewing et al. 1998) bases per sample (not counting failed samples) and a failure rate (<100 Phred 20 bases) of 10% (part of this is attributed to failed or empty template samples).
Considering that there was more than enough signal at 500 nL reaction volume to obtain quality data, further runs were conducted with ½x diluted brews and reaction volumes of 600 nL. Read lengths from one of these runs is shown in Table 2.
Number of Phred 20 Bases/Sample—600 nL Reaction Volume
The average number of Phred 20 bases in the above plate (Table 2) is 547 with a failure rate of 3.1 %. Columns 1–6 were sequenced with undiluted brews and columns 7–12 were sequenced with ½x diluted brews. The average number of Phred 20 bases in the left part of the plate (undiluted) is 556; the average number of Phred 20 bases in the right part of the plate (½x) is 538.
DISCUSSION
The proposed thermal cycling method has been shown to provide rapid thermal cycling of volumes as low as 500 nL to a maximum volume of approximately 5 μL with no significant evaporative loss of sample and in a standard format that is easy to automate. The volume reduction allowed by this method yields a 10-fold or greater reduction in reagent costs for thermal cycling. Based on the present cost of dye terminator reaction brews, the reagent cost per sample with this method can be as little as $0.03 (600 nL reactions with ½ diluted brew). Conversely, undiluted brews might be used to increase cycling reliability with impure templates without paying too high a price for reagents. Initial data indicate that sequence quality obtained using this cycling method with reduced volumes is comparable to that obtained with standard commercially available instrumentation and full reaction volumes.
Although not extensively tested, it is possible that the four separate reactions required for dye primer chemistry can be cycled in a single tip, thus avoiding the four-fold decrease in throughput typically associated with this chemistry.
The reduced amount of product generated by these low-volume reactions is particularly suited for electrokinetic injection in capillary electrophoresis devices which are very sensitive and easily overloaded. Initial results indicate that a 10-fold or greater cost reduction can be achieved with this method without compromising the quality of sequence data. Although at this time the small volume reactions are pipetted into a microtiter plate (and thus diluted) for subsequent cleanup and loading into a capillary sequencer, it is conceivable that the small volume capability and automated pipetting capacity of this technology would make it suitable for interfacing directly to future microfabricated capillary devices.
METHODS
96-Channel Prototype
The 96-channel prototype consists of a 96-channel computer-controlled pipettor on a motorized three-axis gantry operating over a workspace containing a 96-channel cycling block (Fig. 3). The pipettor consists of 96 stainless steel plungers of 0.031″ diameter (Y-SWGX-310, Small Parts) moving inside custom-built syringe bodies (Fig. 1), which are designed to press fit into 30-μL disposable pipette tips (catalog no. 7601, Matrix Technologies). The plungers are actuated by a stepper motor-driven linear stage (Parker 806004CT-D1, 0.2″ lead, 25,000 steps/rev) capable of 0.2-μm resolution. This translates to a minimum aspiration/dispense step of 0.1 nL, which is well beyond the observed reproducibility of the instrument as limited by surface tension effects. A Parker/Hauser XYZ servomotor-driven gantry system is used to position the pipettor over the work area and insert the tips into the block. The work area accessible by the automated pipettor includes new tip boxes, template plates, enzyme brew plates, product plates, lids, a tip melter, a tip cutter, and the thermal cycling block.
Tip melting before insertion into the thermal cycling block is performed by the automated pipettor, momentarily bringing the ends of the pipette tips in contact with a hot plate (held at approximately 200° C) that has been specially modified for increased temperature uniformity.
The temperature of the thermal block is controlled by flowing water through the block and around the tips. Temperature is changed by switching the flow between three different high-flow water baths, each of which is held at one of the temperatures required to perform the denaturing, annealing, and extension stages of thermal cycling, nominally 96°, 50°, and 60° C, respectively. The sealed pipette tips are mounted into the thermal blocks and are surrounded by the high flow, constant-temperature water, which very rapidly brings the reagents inside the tips to the appropriate temperature.
A 96-pin lid is automatically placed onto the pipette tips (using a pneumatic gripper on the pipetting tool head) while the tips are in the thermal blocks to prevent sample evaporation in the blocks during the thermal cycle. The pins on the lid are made of 0.040″-diameter stainless steel pins (SWGX-400-30, Small Parts), completely blocking the top of the pipette tips.
The thermal block is custom-made from acrylic plastic. The top of the thermal block consists of a custom-made aluminum plate, with an O-ring watertight seal to the acrylic housing. The acrylic housing has two intake and two outlet ports, which allow the temperature-regulated water to flow past and surround the pipette tips. Small O-rings are used to seal the pipette tip-to-aluminum plate interface, and a sliding metal plate powered by a pneumatic actuator holds the pipette tips in place when the pipettor disengages from the tips.
Three 28-L water baths (Heating Circulator, Model 8210, Polyscience; Centrifugal Pump, Model ACE-S33, ACE Pump Corp) are used as sources of the three constant temperature fluids used in the thermal cycling sequence. These baths are connected to a PC through serial ports and can be operated by the instrument control software. These baths are further fitted with computer-controlled radiators and fans to allow temperature reduction of the fluid if it exceeds the set-point temperature.
Six water-piloted solenoid valves (VXZ2230, SMC Pneumatics) are used to select the temperature of the water flow entering the thermal block. When a temperature is selected, a pair of input/exhaust valves are opened, and water from the selected bath passes through the thermal block. Calibrated type-T thermocouples (Model 5TC-TT-T-36-36, Omega) with a 0.05 sec time constant are used to monitor the temperatures inside of the temperature bath.
Before dispensing the reaction products into a microtiter plate, the automated pipettor cuts the end of the pipette tips by inserting ∼1/16″ of the ends of all 96 tips through 96 holes in a horizontal guillotine cutter. A pneumatic actuator then forces a blade across the tips, cutting off the melted end.
Control of the pipettor system, gantry, water valves, water baths, and radiators is performed through a PC running LabVIEW (National Instruments) software.
Thermal Cycling Reagents and Conditions
Before aspirating thermal cycling reagents, a mixture of 5 mg/mL BSA (Amersham, 27-8915-01, 9.96 mg/mL stock diluted into dH2O) was aspirated into the 30-μL pipette tips (catalog no. 7601, Matrix Technologies) and ejected, thus coating the interior of the tips with BSA. Subsequently, each tip was loaded with 2 μL of deionized water, a 2-μL air gap, the sample, and finally a second 1-μL air gap. The sample was loaded by aspirating the brew mixture into the tip, and then aspirating the template without aspirating an air gap in between. Template and brew were mixed during the subsequent aspiration of the 1-μL air gap. Brews were Perkin Elmer BigDye Terminators with Taq FS Ready Mix (4303151, PE Biosystems). For a 600 nL reaction, 400 nL of brew were aspirated and then 200 nL of template (40–80 ng/μL).
Undiluted and ½x diluted brews were formed as follows. Undiluted brews: 1 part BigDye Terminator Ready Mix, 1 part Primer (M13-21, 0.8 pmole/μL). ½x brews: 4 parts BigDye Terminator Ready Mix, 8 parts Primer (Operon TGT AAA ACG GCG AGT, 0.8 pmole/μL), 2 parts 5x sequencing buffer (400-mM TrisHCl, 10-mM MgCl2), 1 part dH2O.
The tips were sealed and cycled according to the following protocol: (96° C for 10 sec, 50° C for 5 sec, 60° C for 4 min) × 25 cycles. After cycling, the sample and 2-μL water slug were ejected into 5 μL of deionized water. The resulting product was purified using Sephadex G-50 (DNA Grade F, Pharmacia) by the method described below. Sephadex is washed four times with double volumes of dH2O. The total volume of wash water is eight times the Sephadex volume. 500 μL of the washed Sephadex is loaded into a SilentMonitor filter plate (VWR no. 13586-240). The plate is spun at 2000 g for 2.5 min. Another 300-μL Sephadex is then added and the total is spun again at 2000 g for 2.5 min. The sample is then diluted into 10 μL of dH2O and loaded onto the column, which is spun again at 2000 g for 2 min with a collection plate underneath. This is directly loaded and analyzed on an ABI 3700 using standard running conditions.
Acknowledgments
We thank Scott Hunicke-Smith for his initial work on capillary thermal cycling. We also thank Carl Hansen, Jeff Martin, Azien Safarpour, and Nelson Siu (UBC Department of Physics and Astronomy). This work was funded in part by NSERC and by NHGRI (R01-HG02072-01).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵3 Corresponding author.
-
E-MAIL andre{at}physics.ubc.ca; FAX (604)822-5324.
-
Article published on-line before print: Genome Res.,10.1101/gr.164401.
-
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.164401.
-
- Received September 8, 2000.
- Accepted December 13, 2000.
- Cold Spring Harbor Laboratory Press