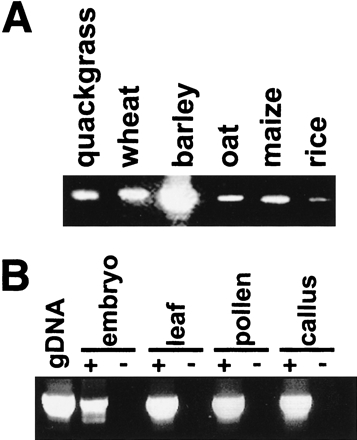
Detection of genomic and expressed env domains. (A) PCR amplification of a 385-bp segment of env domains from 10 ng of genomic DNA of quackgrass (Elymus repens), wheat (Triticum aestivum) barley (Hordeum vulgare), oat (Avena sativa), maize (Zea mays), and rice (Oryza sativa). Bands of identical size could be detected in other Hordeum spp. as well as in rye (Secale cereale). Species other than wheat and quackgrass derive fromPoaceae subfamilies different from that of barley. (B) RT–PCR amplification from RNA of various barley tissues. As a standard, PCR with 10 ng of genomic DNA is shown. The reactions for lanes labeled + contained a reverse transcription step to convert the RNA to cDNA; those labeled − lacked nucleotides at this step as controls for genomic DNA contamination.











