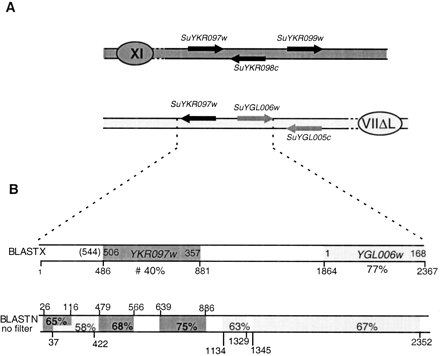
Loss of a duplicated gene in S. cerevisiae outside previously recognized duplication blocks. Symbols are the same as in Fig. 1, and the S. uvarum sequences homologous with S. cerevisiaechromosomes XI and VII are drawn in dark and light gray, respectively. (A) Chromosomal mapping of the two copies ofSuYKR097c in S. uvarum genome. SuYKR097c was isolated twice paired with SuYGL006w and once associated withSuYKR098w and SuYKR099w. (B)BLASTX and BLASTN comparisons (BLOSUM62) between the S. uvarum SuYKR097w-SuYGL006w region and the S. cerevisiaegenome. The sequence of the synteny breakpoint on chromosome VIIΔL was assembled from sequences of overlapping RSTs from the Génolevures projects (XAS0AA01F03TP1, AS0AA19G08DP1, and XAS0AA01F03DP1). Percentage of identity as well as coordinates of the hits are indicated.











