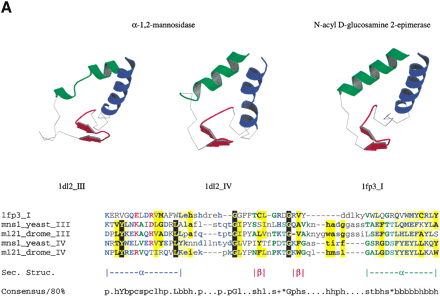
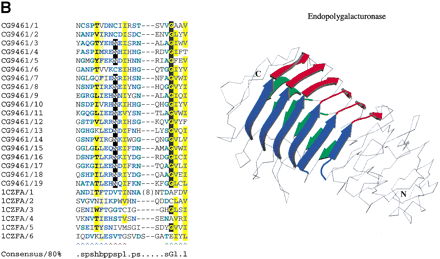
(a) αα repeats found in αα toroids. The third and fourth repeats in yeast α-1,2-mannosidase (PDB code: 1DL2) are detectable by sequence in the equivalent Drosophila protein. The repeating unit is also structurally equivalent to that found in the αα6 class of proteins, such as N-acyl D-glucosamine 2-epimerase (PDB code 1FP3), indicating the two classes of proteins shared an ancestor (see text for details). The sequence alignment produced by aligning the structures of the individual repeats is shown beneath the alignment. Structurally equivalent residues, as defined by the STAMP program (Russell and Barton 1992) are upper case. Nonequivalent regions are lower case. (b) The 3D structure of Aspergillus niger endopolygalacturonase II (PDB code 1CZF). For clarity, the three β-strands in each repeat are colored blue, red, and green. Continuous strands have been split in two at a region corresponding to a pronounced bend in the polypeptide chain, as defined by the DSSP program. Also shown is a multiple sequence alignment of β-helix repeats in A. nigerendopolygalacturonase II (GeneInfo number 6435555) and Drosophila melanogaster CG9461 (GeneInfo number 7299263) represented usingCHROMA (Goodstadt and Ponting 2001). Colored arrows under the alignment correspond to the β-strands in the 3D structure. Asn residues that might form an Asn ladder (see text) are shown as white-on-black. GeneInfo codes for the repeats in PDB:1CZFA andD. melanogaster CG9461 are 6435555 and 7299263; the repeats represent 1CZFA 156–184, 187–206, 209–227, 238–257, 267–287 and 301–327; and CG9461 624–644, 650–670, 673–693, 696–716, 719–739, 742–762, 765–785, 788–808, 811–831, 834–854, 857–877, 880–900, 903–923, 926–946, 949–969, 972–992, 995–1015, 1018–1038, 1041–1061, and 1064–1083.











