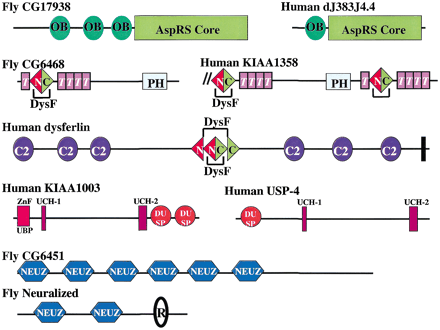
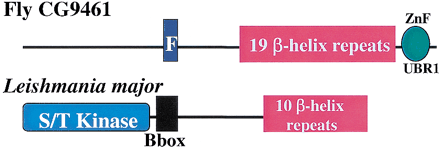
Schematic representation, shown approximately to scale, of the domain architectures of some of the proteins containing repeats or domains that are discussed in the text. Abbreviations: AAA, ATPases associated with a variety of cellular activities; AspRS core, the catalytic portion of aspartyl tRNA synthetases; Bbox, B-box type zinc finger; C2, Protein kinase C conserved region 2 (CalB) domain; F, F-box domain; OB, oligonucleotide/oligosaccharide binding fold; PH, pleckstrin homology domain; R, RING finger domain; T, tectonin-like β-propeller repeats; UCH-1, UCH-2, Ubiquitin carboxyl-terminal hydrolases family 2 (two conserved regions); WD, β-propeller repeat with conserved Trp (W) and Asp (D) residues; ZnF_UBP, Ubiquitin carboxy-terminal hydrolase-like zinc finger; ZnF_UBR1, domain that is involved in recognition of N-end rule substrates in yeast Ubr1p. Solid vertical lines represent predicted transmembrane helices. Double forward slash lines indicate that the protein sequence is N-terminally truncated.











