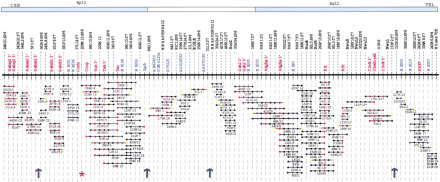
A BAC-based STS/EST-content map of theGabrg1–Gabra2–Gabrb1–Tec–Txk–Pdgfra–Kit–Kdr–Clockregion on mouse Chromosome 5 (oriented with centromeric endleftward and telomeric end rightward). The orthologous regions in the human genome are indicated on thetop (blue and open boxes). The relative positions of mapped STSs and ESTs are indicated on the top of the map, including the corresponding loci names (e.g., D5Buc); the isolated BAC clones are shown as horizontal lines below with circles along the lines indicating positive STS hits. The STSs were developed from BAC insert ends (black); BAC-end STSs corresponding to the original BAC clone (yellow); known genes (red); and SSLP markers (blue). BAC clones were isolated from the C57BL/6J library (RPCI-23). The map is displayed with equal spacing between STS/EST markers and the depicted clones together span a distance of ∼5 Mb. The ordering of five BAC islands in this region was based on the mouse Chromosome 5 genetic linkage, YAC-based, PFGE and RH map (Dietrich et al. 1992; Brunkow et al. 1995; Nagle et al. 1995; King et al. 1997a,b; Tarantino et al. 2000). Purple arrows indicate gaps in the STS-content map, and the red asterisk indicates the gap that was closed by examining the BAC fingerprint cluster shown in Figure 2.











