Partitioning of Tissue Expression Accompanies Multiple Duplications of the Na+/K+ ATPase α Subunit Gene
Abstract
Vertebrate genomes contain multiple copies of related genes that arose through gene duplication. In the past it has been proposed that these duplicated genes were retained because of acquisition of novel beneficial functions. A more recent model, the duplication-degeneration-complementation hypothesis (DDC), posits that the functions of a single gene may become separately allocated among the duplicated genes, rendering both duplicates essential. Thus far, empirical evidence for this model has been limited to theengrailed and sox family of developmental regulators, and it has been unclear whether it may also apply to ubiquitously expressed genes with essential functions for cell survival. Here we describe the cloning of three zebrafish α subunits of the Na(+),K(+)-ATPase and a comprehensive evolutionary analysis of this gene family. The predicted amino acid sequences are extremely well conserved among vertebrates. The evolutionary relationships and the map positions of these genes and of other α-like sequences indicate that both tandem and ploidy duplications contributed to the expansion of this gene family in the teleost lineage. The duplications are accompanied by acquisition of clear functional specialization, consistent with the DDC model of genome evolution.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AY028628, AY028629, and AY028630]
Vertebrate genomes have been shaped by several episodes of large-scale gene duplications. During a very short time in the evolution of early vertebrates, for example, many gene families expanded from one to several paralogs (Sidow 1992; Suga et al. 1999). Such large-scale duplications are the consequence both of individual gene duplications and ploidy duplications, the latter known to have occurred recently in Xenopus and salmonid fishes. There is strong evidence for an older ploidy duplication during the evolution of bony fishes (Postlethwait et al. 1998). For example, seven Hoxclusters are found in zebrafish resulting from a duplication of the four Hox clusters found in most other higher vertebrates (Amores et al., 1998; Prince et al. 1998). As would be predicted from a ploidy duplication, genes linked to the seven Hox clusters are also duplicated with syntenic conservation to the corresponding cluster in mammals (Amores et al. 1998; Woods et al. 2000). Other syntenic regions between human and zebrafish chromosomes lend further support to this model (Postlethwait et al. 2000).
In classical models of genome evolution, the fate of a duplicated gene is dependent upon the nature of the mutations it accumulates. If a new beneficial function is acquired, the duplicate will be retained; if no new function is acquired, it will be lost (Ohno 1970; Sidow 1996). Recently, Force et al. (1999) have proposed an alternate model for certain duplications, one that takes into account the existence of multiple expression domains typically found for products of single genes. Accordingly, the duplication-degeneration-complementation (DDC) model, partial loss-of-function mutations accumulate in thecis regulatory regions of both paralogs, such that each copy of the gene is expressed in a particular spatio-temporal manner, and both copies of the gene must be maintained to preserve the overall function of the original gene. Thus, paradoxically, the accumulation of degenerative mutations enhances the chances of survival of both paralogs. The first example described for the DDC model involves the two engrailed1 genes in zebrafish in which two sites, the pectoral fin buds and spinal cord neurons, express only one of the paralogs (Force et al. 1999). Members of the sox gene family of developmental regulators show a similar partitioning of expression (de Martino et al. 2000). The engrailed and soxduplicate pairs arose from a chromosomal duplication. However, in principle, the nature of the duplication event giving rise to the two paralogs should be irrelevant.
The Na(+),K(+)-ATPase or sodium pump is responsible for maintaining proper intracellular and extracellular concentrations of sodium and potassium ions, and is essential to membrane potential generation (Glynn 1993; Lingrel and Kuntzweiler 1994). The protein consists of a large multi-pass α subunit and an associated glycosylated single-pass β subunit, and has been studied extensively at the biochemical level. The highly conserved α family of genes that encode the catalytic subunits of the pump contain the binding sites for Na(+), K(+), ATP and the digitalis glycosides (Lingrel and Kuntzweiler 1994). The mature protein can be associated with a third, smaller γ subunit (Mercer et al. 1993). Invertebrate genomes possess one α subunit of the Na(+),K(+)-ATPase (Emery et al. 1995). Four α subunits have been reported in mammalian systems (Kawakami et al. 1986; Shull et al. 1986;Martin-Vasallo et al. 1989; Shamraj and Lingrel 1994; Malik et al. 1996). In mammals, the expression patterns of the different genes show strong tissue preferences (Sweadner 1989; Blanco and Mercer 1998). Theα1 gene (atp1a1) is preferentially expressed in the kidney, gut, and heart, and ubiquitously at lower levels. Theα2 (atp1a2) gene is expressed in muscle, adipocytes, heart, and brain. The α3 (atp1a3) gene is expressed throughout the nervous system. A less conserved fourth isoform, α4, is expressed in the testes (Shamraj and Lingrel 1994). The α1 isoform is essential to cell survival. Homozygous disruption of this gene in mice causes embryonic lethality (James et al. 1999).
We evaluated these genes in the zebrafish because of evidence that it has undergone an additional round of genome duplication over 100 million years ago (Amores et al., 1998). Through cloning and database searches, we discovered that zebrafish have at least eight α subunits. Five are in the α1 class, two in the α3class, and one in α2 class. Sequence, mapping, and expression analyses all suggest that even in the relatively short period of time since gene duplication, they have acquired specialized functions, supporting the subfunctionalization model of genome evolution.
RESULTS
Homology to Other α Subunits
We cloned three α subunits of the Na(+),K(+)-ATPase. Two of the genes belong to the α1 family and the third is in theα2 group. From the predicted amino acid sequences, the twoα1 genes share 91% identity between each other and 83% identity with the α2 homolog (Fig.1). This amino acid identity is preserved when comparing the zebrafish subunits with their human counterparts.atpα1A1 and atpα1B1 show 88% and 89% identity, respectively, to the human α1 subunit and atpα2 shares 86% identity to the human α2 protein.
Predicted amino acid alignment of the three zebrafish sodium pump α subunits. Identities are shown in black boxes, similarities in gray. The TGES motif is underlined and the phosphorylated aspartate is shown by a triangle. The conserved PKA phosphorylation site is marked by an asterisk. The broken line indicates the ankyrin repeat binding site.
A number of motifs essential for the sodium pump's function are noted in Figure 1. The conserved cytoplasmic TGES/A motif, critical to the catalytic cycle of all P-type ATPases, is present in all three of the zebrafish α subunits. The catalytically phosphorylated aspartyl residue is also conserved and found within the consensus DKTGTLTQ sequence. Protein kinases can modulate the catalytic activity of the sodium pump in a complex fashion (Therien and Blostein 2000). Phosphorylation by PKA is thought to occur at Ser 943 of the human and rat sequences. This serine is also found in all three of the zebrafish isoforms within the context of a PKA consensus site (XRRXSX). The Na(+),K(+)-ATPase can also bind to ankyrin repeats (Zhang et al. 1998), and this binding site is conserved in all three fish isoforms. The presence of a leucine residue in the α1 genes instead of a methionine is unusual, although it is found in the related P-type pump, the gastric H(+),K(+)-ATPase.
Ouabain resistance in the rat α1 isoform has been mapped to the extracellular loop between the first and second membrane-spanning segments. Mutation of two amino acid residues within this loop to those of rat α1 can confer resistance to ouabain-sensitive isoforms (Price et al. 1990; Jewell and Lingrel 1992). The zebrafish genes have sequences predicted to be ouabain sensitive in that they contain a glutamine or leucine rather than an arginine residue found at the first position of the resistant isoform, and an asparigine instead of an aspartate at the second.
Phylogenetic Analysis
To generate an evolutionary comparison, we identified additional vertebrate α genes in GenBank. Between gene cloning and database analysis, we have identified a total of eight αgenes. Five are α1 paralogs, two are α3 paralogs, and one is an α2 gene. (The α4 gene in mammals is far less conserved and sequence information from multiple species is lacking, so we did not include it in the analysis.) Protein maximum likelihood analysis predicts the evolutionary tree shown in Figure2.
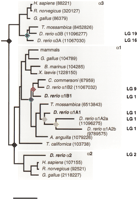
Phylogenetic analysis of vertebrate sodium pump catalytic subunits. Invertebrate sequences are used to root the tree. Gene duplication events are indicated by colored rhomboids. The tree is drawn to scale with the horizontal distances joining two nodes proportional to the number of amino acids substitutions. Scale bar, 10% substitution. The GenPept protein identification numbers for each gene are given. Genes in boldface represent those described in the present study. Partial sequences were not used in the analysis. For clarity, α1 sequences for mammals are grouped into one branch because the number of amino acid substitutions are small. The mammalian sequences includedEquus caballus (89128), Canis familiaris (1703466),Ovis aries (67958), Rattus norvegicus (92517),Homo sapiens (88220), and Sus scrofa (89248). Chromosome assignments for the zebrafish subunits were obtained by radiation hybrid mapping and are indicated at right.
Invertebrate genomes contain only one α subunit (Emery et al. 1995), which we used to root the tree. The first gene duplication created the α3 branch and an ancestral α1α2 gene, which then duplicated to give rise to the α1 and α2paralogs. Nodes that correspond to these duplications are shown in black in Figure 2, and are likely due to the large-scale gene duplications that occurred in the ancestral lineage of vertebrates. All additional gene duplications appear only in the teleosts. The duplication giving rise to the two α3 paralogs (atpα3A and atpα3B) appears to have preceded the divergence of the Tilapia and zebrafish lineages (ProtMLBootP value = 83%) and is likely to have occurred as part of the known large-scale gene duplication in the teleost lineage.
The α1 gene appears to have undergone additional rounds of duplication. The first α1 gene duplication event appears to have occurred in an early bony fish ancestor prior to the divergence of the lineage leading to present day eels (Fig. 2, blue node). Placement of the eel lineage before the gene duplication is rejected at a ProtMLBootP value of 0.001%, confirming that this duplication was not due to the known large-scale teleost duplication. The two resultingα1 homologs then further duplicated, generating at least five α1 genes in zebrafish. Duplication of theatpα1A group (gray node) occurred after divergence from theTilapia lineage (supported at BootP = 100%). A subsequent duplication (yellow node) generated the other two knownatpα1A homologs (atpα1A2a andatpα1A2b). The one gene duplication in the subtree containing atpα1B1 (red node) occurred before the divergence of the lineage leading to the sucker fish (Catostomus commersoni), suggesting that this duplication may have been due to the known large-scale teleost genome duplication.
Map Positions of the Zebrafish Catalytic Subunits
To examine the origin of the duplications, we mapped all of these genes by somatic cell radiation hybrid (RH) mapping (Geisler et al. 1999). As shown in Figure 2, atpα2 is on linkage group 2 (LG 2), whereas atpα1A1 and atpα1B1 both map to LG 1 in the z9394–z9382 interval. The atpα3A gene maps to LG 16 close to the Hoxab cluster, and atpα3B maps to LG 19 near the Hoxaa cluster. Both atpα1A2a andatpα1A2b map to LG 1 in the same interval asatpα1A1 and atpα1B1, whereas atpα1B2maps to LG 9 between markers z9112 and z20031 (Shimoda et al. 1999).
Expression of Zebrafish α1 Isoforms
If the DDC model of subfunctionalization of genes after duplication applies, expression patterns should have been partitioned among the zebrafish ATPases relative to, for example, the single mammalianα1 gene. Therefore, we examined the expression ofatpα1A1 and atpα1B1.
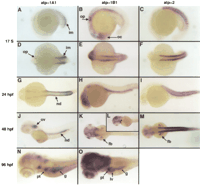
The expression patterns of atpα1A1 and atpα1B1 in early development are quite distinct. At the 17 somite stage,atpα1A1 is expressed in the intermediate mesoderm and, at lower levels, in the otic placode (Fig.3A,D), whereas the atpα1B1 gene is expressed in the optic cup, the developing nervous system, the otic placode, and, weakly, in the intermediate mesoderm (Fig. 3B,E). Theatpα2 gene is expressed in the developing somites (Fig.3C,F). Thus, at this early stage, atpα1A1 is the dominant kidney isoform, whereas atpα1B1 is expressed in a variety of other tissues.
Whole-mount expression of atpα1A1, atpα1B1 andatpα2 during zebrafish development. Parallel expression analysis of zebrafish sodium pump catalytic subunits at the 17 somite, 24 hpf, 48 hpf, and 96 hpf stages. At the 17 somite stage,atpα1A1 is expressed in the intermediate mesoderm and at lower levels in the otic placode (A,D);atpα1B1 is expressed in the otic placode, optic cup and throughout the brain (B,E); atpα2 is expressed in the somites (C,F). At 24 hpf,atpα1A1 expression is seen in the nephric duct (G) and lower levels in the otic vesicle; atpα1B1 is expressed in the brain and nephric duct (H), whereas atpα2expression remains exclusive to the muscle lineage (I). At 48 hpf, atpα1A1 expression is detected in the otic vesicle and nephric duct (J); atpα1B1 expression remains in the brain, but is now also seen in the fin buds (K) and the heart (asterisk in L); atpα2 is also detected in the fin buds (M). (N,O) Expression ofatpα1A1 and atpα1B1 at 96 hpf, note the absence of atpα1A1 expression in the liver and the strong expression of atpα1B1 in the brain. (im) Intermediate mesoderm; (op) otic placode; (oc) optic cup; (nd) nephric duct; (ov) otic vesicle; (fb) fin bud; (pt) pronephric tubule; (g) gut; (lv) liver.
By 24 h postfertilization (hpf), atpα1A1 is still expressed in the nephric duct (Fig. 3G) and, to a lesser extent, in the otic vesicle. We also detect weak expression of atpα1B1 in the kidney, but it remains expressed in many tissues (Fig. 3H). Expression of atpα2 remains exclusive to the myogenic lineage (Fig.3I). At 48 hpf, expression of atpα1A1 in the ear has increased and is comparable with the levels seen in the nephric duct (Fig. 3J). The dominant isoform in the nervous system isatpα1B1 (Fig. 3K). It is also transiently expressed in the pectoral fin buds (Fig. 3K) and the heart, in which the ventricular myocardium expresses much higher levels of atpα1B1, than does the atrium (Fig. 3L). The atpα2 gene remains in the somites and is also seen in the fin buds. (Fig. 3M).
At 96 hpf, atpα1B1 is highly expressed in the brain (Fig.3O). Low levels of atpα1A1 can also be detected in this tissue (Fig. 3N). Both genes are expressed in the pronephric tubule and gut to a similar level (Fig. 3N,O). However, only theatpα1B1 paralog is detected in the developing liver (Fig. 3, cf. N and O).
DISCUSSION
Tandem and Ploidy Gene Duplications
Invertebrate genomes contain one sodium pump catalytic subunit (Emery et al. 1995). We show here that the three vertebrate paralogs originated by gene duplications that preceded the last common ancestor of jawed vertebrates (which is represented by the node in the α1 subtree from which the lineage to Torpedo californicaoriginated; Fig. 2). We find that the zebrafish genome encodes at least eight subunits that fall into the α1, α2, orα3 subfamilies. Our phylogenetic analysis indicates that a gene duplication of the α1 gene took place in the bony fish lineage, creating the α1A and α1B subfamilies. Further duplications within each of the branches generated the additional α1 genes found in the zebrafish. Whether this additional expansion is unique to zebrafish awaits further genomic analysis of other fish.
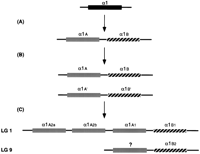
Combining data from the phylogenetic analysis and mapping, a clear picture of the gene duplication events in teleosts emerges (Fig.4). The first duplication, which created the α1A and α1B subfamilies, was likely a tandem duplication because (1) it preceded the known large-scale duplication, which occurred after divergence of the lineage leading to eels, and (2)α1A and α1B both map to the same small interval on LG 1 (Fig. 2).
Diagram illustrating the gene duplication events for the α1gene in zebrafish. (A) A tandem duplication generated twoα1 paralogs as follows: α1A, shown in gray andα1B, hashed. (B) The tandem is duplicated by the large scale duplication in the bony fish lineage α1A andα1B. (C) Present-day zebrafish. One duplicate pair is found on LG 1, where the α1A paralog has undergone two further gene duplications to generate three genes (α1A1,α1A2a, and α1A2b). We do not have evidence for the existence of the other α1A paralog. It may have been lost or remained as a pseudogene and is indicated by a question mark. The α1B paralogs have give rise to the α1B1 gene on LG 1 and α1B2 on LG 9. The arrangement of the genes on the chromosome is arbitrary.
The duplication giving rise to the two α3 paralogs and the duplication in the α1B subfamily are likely due to the teleost large-scale duplication. Both the linkage data and the phylogenetic analysis favor this interpretation. The two α3paralogs map to LG 16 and LG 19, linked to the duplicated Hoxaclusters (Amores et al. 1998; Gates et al. 1999; Woods et al. 2000), and the duplication occurred prior to divergence of the lineage leading to Tilapia. Similarly, the α1B paralogs map to LG 1 and LG 9, which have been suggested to contain paralogous regions, as teleost-specific members of other gene families, such asdistalless and engrailed, map to these chromosomes (Gates et al. 1999; Woods et al. 2000). Here too, the phylogenetic position of the duplications is consistent with the large-scale duplication. Duplicates of the atpα1A lineage are unlikely to be due to the large scale gene duplication, as both duplications follow the species divergence node with Tilapia. Radiation hybrid mapping of all three genes indicates that they lie in the same area of LG 1, suggesting that they arose by tandem duplication.
Divergence of Expression and the DDC Model
Gene duplications have been proposed to be an important mechanism driving diversification during evolution. According to classical models, duplicated genes can accumulate mutations and acquire novel functions. Alternatively, they may be lost, such as one of theHoxd clusters in zebrafish, or may degenerate to a pseudogene, as Hoxaa-10 has in zebrafish (Stellwag 1999). According to the DDC model (Force et al. 1999), some degenerative mutations may actually favor the preservation of both paralogs through complementation of their subfunctions.
Thus far, evidence for the DDC model has come from genes encoding developmental regulators. The zebrafish genome contains twoengrailed1 genes expressed in two distinct locales of a few cells each, in the fin buds and spinal cord (Force et al., 1999). The expression pattern of the duplicated sox11 genes in zebrafish is also partitioned. Whereas the single sox11 gene is expressed throughout the somites in the mouse, two zebrafish paralogs share this expression domain; sox11a is expressed anteriorly and sox11b posteriorly (de Martino et al., 2000). Here we present data that extend this model to genes essential for survival of every cell in the body. Interestingly, data from studies of the tissue-specific expression of isozymes of lactate dehydrogenase and alcohol dehydrogenase (Li et al. 1983; Edenberg 2000) are also consistent with a DDC mechanism; future genetic mapping and evolutionary anlyses of these genes may provide further insights into the molecular mechanisms of DDC.
The DDC model predicts that duplicate genes partition functions normally performed by the original gene. Our expression results foratpα1A1 and atpα1B1 are consistent with this model. The α1 isoform in mice is expressed in the kidney, heart, brain, and gut. In zebrafish, the atpα1A1 gene is the dominant kidney isoform expressed very early in nephrogenesis and at very high levels. The atpα1B1 gene is highly expressed in the brain. The other α1 paralog (atpα1A1) is also expressed in the brain, but much later in development and at lower levels. Only atpα1B1 expression is detected in the heart and liver. The model further predicts that the broader the expression domains associated with a gene, the more likely duplicated paralogs will be maintained. The α1 gene in mammals is ubiquitously expressed, performing essential functions in all cell types of the organism. In other words, it contains the greatest potential number of subfunctions. Consistent with this prediction, the α1 family in zebrafish contains the greatest number of paralogs with five.
Prior data for the engrailed1 genes indicate that partitioning of expression domains can follow a chromosomal duplication event. Our mapping, expression, and evolutionary data for the α family members are consistent with these prior observations and reveal, as well, examples of subfunctionalization following a tandem duplication event. These data lend further support to the DDC model, extending it to ubiquitously expressed genes and to tandem duplications.
METHODS
Zebrafish Lines
Fish were raised and maintained in the Cardiovascular Research Center fish facility at the Massachusetts General Hospital. Embryos were kept in E3 medium at 28.5°C and staged according to somite number or hours postfertilization (hpf) (Kimmel et al. 1995). Wild-type fish were of the Tübingen background (obtained from Dr. Christianne Nüsslein-Volhard, Tübingen).
cDNA Cloning and Sequence Analysis
We identified an expressed sequence tag (accession no. AA494679) encoding a zebrafish α subunit in the public databases and PCR primers were designed to amplify a portion of this coding sequence. We used the PCR product to screen 5 × 106 plaques at low stringency from a 24-h zebrafish cDNA λZAP Express library (Stratagene). We selected 20 plaques for further study of ∼300 positives obtained. Ten strong positives and ten weaker positives were selected and rescued. Full-length clones were completely sequenced on both strands and characterized. We retrieved homologs from GenBank byBLAST search, and aligned the amino acid sequences withCLUSTALW (Thompson et al. 1994). Regions of uncertain homology around gaps were excluded from subsequent phylogenetic analysis by use of PROTML (Adachi and Hasegawa 1996). The tree was rooted with invertebrate sequences that were later excluded from the analyses of the vertebrate sequences to maximize the number of unambiguously homologous sequence positions. The topologies of α1, α2, and α3 subtrees were determined in independent analyses, and a global analysis allowed determination of the relative branching order of the three paralogs. Branching order in those parts of the tree that are important for our interpretation of the nature of the nodes was tested explicitly by use of the user tree option inPROTML.
In Situ Hybridizations
Embryos of the appropriate stage were fixed in 4% paraformaldehyde, stored in methanol and processed as described previously (Jowett 1999). The cDNA clones (in pBK-CMV) were linearized with BamHI and T7 RNA polymerase used to transcribe antisense DIG-labeled riboprobes.
Radiation Hybrid Mapping and Genotyping
We designed primers to amplify a portion of the 3′ untranslated region of each α subunit. We also mapped GenBank entries representing other α-like genes. In Table 1, we show our proposed nomenclature consistent with the phylogenetic data presented in this study.
Primer Pairs Used in Radiation Hybrid Mapping
The cycling profile was as follows: 90 sec of denaturation at 94°C, 30 cycles with 30 sec at 94°C, 30 sec at 60°C, and 1 min at 72°C. Somatic cell radiation hybrid mapping was carried out by use of the Goodfellow T51 panel (Research Genetics) and map positions calculated by use of the RH mapping service at Boston's Children's Hospital (http://genetics.med.harvard.edu/∼zonlab/).
Acknowledgments
We thank Alex Therien for critical comments on earlier versions of this manuscript and for helpful discussions and Sarah Childs for helpful criticism of the manuscript. This work was supported in part by National Institutes of Health grants RO1DK55383 and RO1HL63206 to M.C.F.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
An analysis of zebrafish α subunit genes has also been performed by Rajarao et al. (2001).
Footnotes
-
↵3 Corresponding author.
-
E-MAIL fishman{at}cvrc.mgh.harvard.edu; FAX (617) 726-5806.
-
Article published on-line before print: Genome Res., 10.1101/gr.192001.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.192001.
-
- Received April 12, 2001.
- Accepted June 4, 2001.
- Cold Spring Harbor Laboratory Press