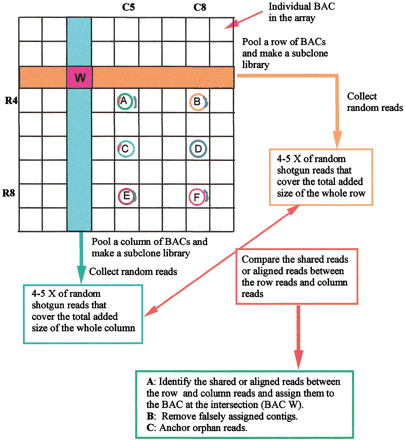
General Clone-Array Pooled Shotgun Sequencing (CAPSS) strategy. Genomic clones (e.g., BACs) are organized in a two-dimensional array, and pools of DNA from each row and column are converted to a subclone library for sequencing. The sequence assembly of each clone is generated by cross assembly of each row and column, shown as clone W in this schema. Clones A–F exemplify possible complications from other overlapping sequences in the array. The colors in clones A–F represent unique sequences. Here clone A and clone C share sequence, as do B and E. Cross assembly of R4 + C5 will yield assembly from BAC A, and will include reads from the overlap in BAC C. Clones B and E will also generate contigs from both assemblies of R4 + C5 and R8 + C8. The generation of contigs at multiple locations in the grid distinguishes overlap that does not originate from the clone at the row/column intersection. Circles in A–F represent a perfect complete sequence contig, with colors coding for different sequences. Note that the shared sequence contig (in blue) between clone B and clone E will lead to assignment of the same contig to clone A and F, shown as an independent contig (in blue).











