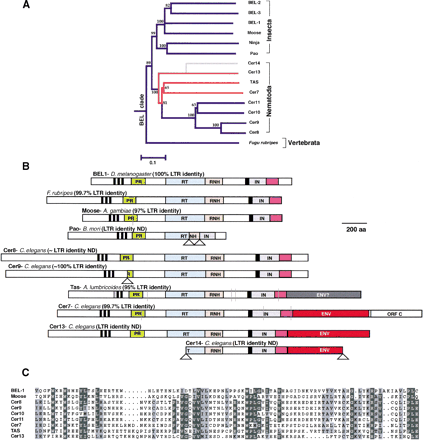
(A) Phylogenetic analysis of BEL clade members from insect, nematode, and vertebrate genomes. Bootstrap values and a divergence scale are indicated. (B) Schematic ORFs from representative members of the BEL clade with various enzymatic and structural features highlighted. Vertical gray lines indicate a termination codon or frameshift encountered. Lower triangles indicate larger deletions. Two different env genes are found in Tas and the Cer7, Cer13, and Cer14 retroviruses (Bowen and McDonald 1999; Felder et al. 1994). In Cer7, an additional accessory protein is found downstream from theenv gene (Bowen and McDonald 1999). A carboxyl-terminal extension to the core integrase domain, with a presumed DNA-binding role is also highlighted, and a multiple alignment presented in (C). There has been some confusion over the enzymatic domains encoded by the BEL clade based on the apparent absence of an intact ribonuclease H/integrase domain from one of the earliest members identified, Pao (B. mori). Closer inspection from pairwise comparisons reveals that this is the result of at least two large internal deletions in the open reading frame of the Pao element that was originally sequenced (Xiong et al. 1993). This is confirmed from comparisons to intact Pao elements from the B. mori genome whose sequence is present in the est (expressed sequence tags) databases.











