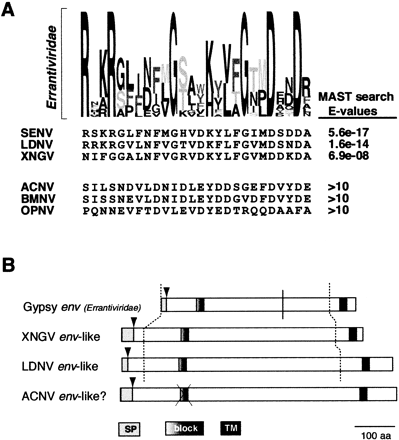
(A) Logos of the conserved block in the envelope genes of insect errantiviruses. In the Logos format, the height of each residue is proportional to its frequency, and the total height of all the residues in the position are proportional to the conservation (information content) at any particular position. Thus, the tallest residues represent invariant residues. This information is used to construct weighted queries to search the protein database. Highlighted below the Logo are the significant MAST matches and the E-values reported. Because blocks are ungapped, a gap was manually introduced in the gypsy sequence to correct obvious misalignments (Fig. 2). (B) Schematic ORFs of the env-related genes in errantiviruses and baculoviruses. Highlighted are the predicted leader peptide (SP, cleavage site shown by the arrow) and transmembrane regions (see Methods), as well as the conserved block of conservation (Fig. 2) common to all errantiviruses. The solid vertical line indicates the site of proteolytic cleavage for the gypsy env, whereas the dotted lines refer to the region aligned in Fig. 2.











