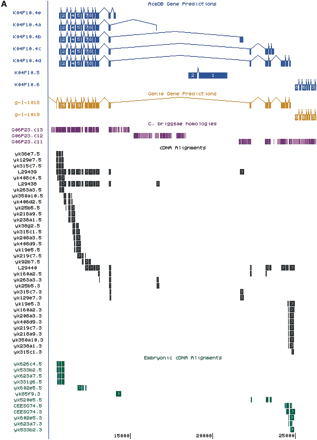
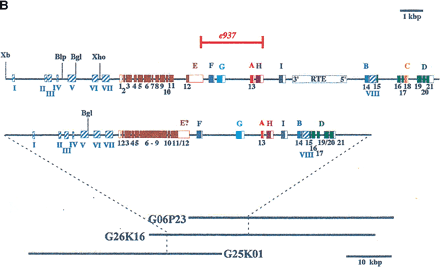
(A) Exon prediction in genomic regions near bli-4(also called K04F10; from Kent and Zahler athttp://www.cse.ucsc.edu/∼kent/cgi-bin/tracks.exe?where=K04F10). (B) Schematic representation of the bli-4 gene fromC. elegans (top) and C. briggsae(bottom). The common region(s) encoding the signal peptide, prodomain, protease domain, and middle domain are shown in brown. Alternatively spliced exons that encode carboxyl termini unique to the individual isoforms are labeled alphabetically and color coded. The position and extent of the e937 3325-bp deletion is indicated. Open boxes represent noncoding exons or untranslated regions; shaded boxes represent coding exons. Hatched boxes (labeled I–VIII) represent regions of nucleotide homology that may constitute regulatory elements, particularly those at the 5′ end. (Xb) XbaI; (Blp) BlpI; (Bgl) BglII; (Xho) XhoI. The approximate location of the C. briggsae bli-4 gene is indicated by dashed lines on the fosmid clones used in Thacker et al. (1999). The entire sequence of clones G25K01 and G06P23 has been determined, whereas G26K16 was used solely for transformation rescue experiments. (Reprinted with permission from Thacker et al. 1999.)











