“Spot-On” SNP Genotyping
As we enter the post-genomic era, where complete reference genome sequences are a given, analyzing the variation among individual genomes is an area of intense investigation. One of the most common forms of genomic variation is single nucleotide polymorphisms, or SNPs. The “science of SNPs” has become a heavy industry, as evidenced by the proliferation of methods for genotyping and deployment of entire commercial enterprises devoted to research centered on SNPs. Why are we seeing such keen interest in these technologies?
Applications that utilize SNPs are cropping up in diverse areas of research. Undoubtedly, the area where SNPs are receiving the most attention is their use as genetic markers for the study of complex human traits and pharmacogenomics. Genome-wide complements of SNPs are now being developed as genetic tools in model organisms such asArabidopsis, Drosophila, and C. elegans. SNPs are finding favor as markers for genetic crosses and selective breeding strategies in livestock and plants of agricultural importance. All of these applications demand a technology that provides simple, robust, highly accurate and cost-effective genotyping of large numbers of SNPs on hundreds or thousands of individuals. Unlike DNA sequencing, where separation-based dideoxy sequencing predominates, no single technology for scoring SNPs has become a widely accepted standard. In this issue, Pastinen and colleagues present a novel array-based method for SNP genotyping (Pastinen et al. 2000).
Array of Light
The genotyping system developed by groups at the National Public Health Institute of Finland and Uppsala University uses allele-specific oligonucleotides on microarrays as templates for primer extension-based genotyping (Figure 1A). The technique, derived from earlier work of the same group (Pastinen et al. 1997), has several features that facilitate high-throughput genotyping. In order to extract the genomic sequence harboring the SNP, Pastinen and colleagues developed a two-step process whereby the sequence is first amplified via the PCR with T7-tailed primers and then transcribed into RNA by T7 RNA polymerase. These RNA molecules serve as templates for primer extension with reverse transcriptase. The process includes very specific conditions that allow the T7 and RT reactions to be performed in a single step directly on the arrays, greatly reducing the number of manipulations required. Furthermore, the group presents data demonstrating that allelic discrimination power is retained when highly multiplexed templates are used, which permits genotyping many SNPs simultaneously on a single array.
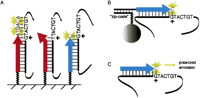
Recent approaches to primer extension-based genotyping. A hypothetical GT polymorphism and flanking 3′ sequence is shown in these examples. Primers used for extension reactions are represented by large arrows, PCR-amplified genomic DNA containing the SNP as solid black lines, and linker molecules as zigzag lines. Small arrows indicate the SNP being assayed, fluorescently labeled nucleotides are in red, and green flashes represent fluorescent emission in response to appropriate excitation light. (A) Using microarrays, the extension primer is anchored to an array surface, and the extension occurs either in an allele-specific manner, as shown by the leftmost red arrow, where the 3′ base of the primer is complimentary to one allele of the variant base. This primer does not hybridize to the other allele and thus does not extend (middle red arrow, Pastinen et al. 2000). Another approach is to design the extension primer (blue arrow) to end just 5′ of the SNP such that discrimination depends on the incorporation of a fluorescently labeled dideoxynucleotide, in this case C (Pastinen et al. 1997, Kurg et al. 2000). The identity of the incorporated base can also be determined without fluorescence using mass spectrometry (Ross et al. 1998, Tang et al. 1999). (B) In one bead-based solid phase approach, generic “zip-code” oligonucleotides are anchored to labeled microspheres and specifically “capture” the complimentary sequence linked to the extension primer. Discrimination is achieved by FACS sorting the microspheres and determining captured fluorescence due to the incorporated nucleotide (Chen et al. 2000). Approaches where the primer is biotinylated have also proven useful. Generic “zip-code” oligonucleotides have also been used on arrays (Gunderson et al. 1998, Favis et al. 2000). (C) Using fluorescence polarization, allelic discrimination can be achieved without a separation step by assaying nucleotide-specific polarized emission in response to polarized excitation light (Chen et al. 1999).
A critical consideration when evaluating any new genotyping technology is how well it performs in an actual application to a large set of SNPs. Pastinen et al. demonstrate the accuracy and robustness of their system by generating ∼8,000 genotypes on a collection of 40 known mutations and SNPs. They further illustrate the sensitivity of the assay in detecting alleles that are diluted to low copy number. This suggests that their system may have utility in genotyping approaches requiring detection of rare alleles, such as pooling strategies for detection of identical-by-descent regions, or detection of somatic mutations in tumor samples.
How does the Pastinen group's system compare to other genotyping technologies? Microarrays are an attractive platform for SNP analysis, notably because of the capacity for interrogating a large number of SNPs simultaneously on a single array. Previous reports demonstrating the use of microarrays in SNP genotyping employed allele-specific hybridization as the discrimination method (Wang et al. 1998; Hacia et al. 1999). Because of the highly redundant and complex nature of the arrays required, the utility of this approach has been limited to a few select groups with access to sophisticated manufacturing and probe design capabilities. In contrast, the array system used by Pastinen et al. relies on a simple arraying robot of the type that has found regular use at many institutions for constructing gene expression arrays. In addition, the chemistry for anchoring the oligonucleotides to the arrays is one commonly used. This type of array offers flexibility, allowing the investigator to redesign probes and arrays as differing needs arise without a heavy up-front investment in probe design or array manufacturing.
Enzymatic Assistance
Pastinen et al.'s technique relies on allele-specific primer extension with reverse transcriptase. This combination of sequence-specificity (the allele-specific primers) and the added signal strength offered by the enzyme is powerful, and is a common feature of several genotyping systems. One approach uses DNA ligase, in which ligation of two allele-specific oligonucleotides occurs in a sequence-dependent manner. This method has been adapted to both solid phase and array based genotyping (Nickerson et al. 1990; Gunderson et al. 1998; Iannone et al. 2000). Allele-specific, enzymatic cleavage of “flap“ DNA probes has also been shown to be an effective method (Lyamichev et al. 1999). Perhaps because of parallels with methods for DNA sequencing, a number of methods using a primer extension reaction for genotyping have recently appeared. Most of these approaches can be classified as either requiring allele-specific primers with deoxynucleotide incorporation, or non-allele-specific primers and incorporation of dideoxynucleotides, as is done in DNA sequencing. Many of these methods rely on fluorescently labeled nucleotides or probes, and thus one of the challenges is how to efficiently separate the incorporated nucleotides (signal) from the unincorporated (noise). As shown in Figure 1, one approach is to couple the extension primer to a solid phase, such as an array surface or bead. Beads can be selected out based on either affinity or flourescent properties (Nikiforov et al. 1994; Chen et al. 2000), and the identity of the incorporated base can be determined either by flourescent labels or mass spectrometry (Ross et al. 1998; Tang et al. 1999).Two approaches that eliminate the need for a separation step, FRET-TDI and FP-TDI, instead make use of the inherent properties or interactions of fluorescent molecules in the solution to distinguish the signal (Figure 1C) (Chen and Kwok 1999; Chen et al. 1999).
Looking Forward
Pastinen and colleagues demonstrate the tremendous potential for SNP genotyping on arrays. Five thousand genotypes are currently possible on a single slide, and 20,000 genotypes per slide will be possible with minor improvements in spotting density. This throughput is comparable to most high-capacity genotyping systems currently available, with the added benefit of a flexible array format. What are some remaining issues? First, developing efficient strategies for accessing the genomic sequence harboring each SNP remains a challenge, because current multiplex PCR strategies are difficult to optimize and apply on a large scale. Second, standardized arraying protocols need to be developed, because the spot quality of each array can vary and thus dramatically affect the quality of the data. Third, developing a sensitive four-color detection system will permit alternate primer extension approaches. Such a system is already in development from at least one group (A.-C. Syvänen, pers. comm.). Finally, as with gene expression data from microarrays, the need for powerful software to interpret and organize the large amounts of data will be paramount to further development.
How does one choose among the multitude of SNP genotyping technologies? All technologies have attractive advantages. The individual investigator has to evaluate his or her own needs for throughput, cost, and experimental goals in order to make an informed decision. Looking ahead, novel array formats are emerging (Dubiley et al. 1999; Favis et al. 2000), in some cases with substantial commercial backing; no doubt other assay formats are in development. At the moment, no single technology has monopolized the field and it seems unlikely that we'll be seeing an antitrust investigation from the Justice Department anytime soon!
Footnotes
-
↵1 E-MAIL ; FAX 650 837-8240.
- Cold Spring Harbor Laboratory Press