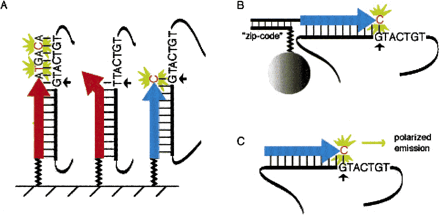
Recent approaches to primer extension-based genotyping. A hypothetical GT polymorphism and flanking 3′ sequence is shown in these examples. Primers used for extension reactions are represented by large arrows, PCR-amplified genomic DNA containing the SNP as solid black lines, and linker molecules as zigzag lines. Small arrows indicate the SNP being assayed, fluorescently labeled nucleotides are in red, and green flashes represent fluorescent emission in response to appropriate excitation light. (A) Using microarrays, the extension primer is anchored to an array surface, and the extension occurs either in an allele-specific manner, as shown by the leftmost red arrow, where the 3′ base of the primer is complimentary to one allele of the variant base. This primer does not hybridize to the other allele and thus does not extend (middle red arrow, Pastinen et al. 2000). Another approach is to design the extension primer (blue arrow) to end just 5′ of the SNP such that discrimination depends on the incorporation of a fluorescently labeled dideoxynucleotide, in this case C (Pastinen et al. 1997, Kurg et al. 2000). The identity of the incorporated base can also be determined without fluorescence using mass spectrometry (Ross et al. 1998, Tang et al. 1999). (B) In one bead-based solid phase approach, generic “zip-code” oligonucleotides are anchored to labeled microspheres and specifically “capture” the complimentary sequence linked to the extension primer. Discrimination is achieved by FACS sorting the microspheres and determining captured fluorescence due to the incorporated nucleotide (Chen et al. 2000). Approaches where the primer is biotinylated have also proven useful. Generic “zip-code” oligonucleotides have also been used on arrays (Gunderson et al. 1998, Favis et al. 2000). (C) Using fluorescence polarization, allelic discrimination can be achieved without a separation step by assaying nucleotide-specific polarized emission in response to polarized excitation light (Chen et al. 1999).











