Alu Elements Support Independent Origin of Prosimian, Platyrrhine, and Catarrhine Mhc-DRB Genes
Abstract
The primate major histocompatibility complex (Mhc) genes fall into two classes and each of the classes into several families. Of the class II families, the DRB family has a long and complex evolutionary history marked by gene turnover, rearrangement, and molecular convergence. Because the history is not easily decipherable from sequences alone, Alu element insertions were used as cladistic markers to support the surmised phylogenetic relationships among the DRB genes. Intron 1 segments of 24 DRBgenes from five platyrrhine species and five DRB genes from three prosimian species were amplified by PCR and cloned, and the amplification products were sequenced or PCR–typed for Alurepeats. Three Alu elements were identified in the platyrrhine and four in the prosimian DRB genes. One of the platyrrhine elements (Alu50J) is also found in the Catarrhini, whereas the other two (Alu62Sc, Alu63Sc) are restricted to the New World monkeys. Similarly, the four prosimian elements are found only in this taxon. This distribution of Alu elements is consistent with the phylogeny of the DRB genes as determined from their intron 1 sequences in an earlier and the present study. It contradicts the exon 2-based phylogeny and thus corroborates the conclusion that the evolution of DRB exon 2 sequences is, to some extent, shaped by molecular convergence. Taken together, the data indicate that each of the assemblages of DRB genes in prosimians, platyrrhines, and catarrhines is derived from a separate ancestral gene.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos.AF197226–AF197240.]
The major histocompatibility complex (Mhc) is a multicomponent assemblage comprised of genes of different age (Parham 1999). All jawed vertebrates possess two classes of Mhc loci and in each class there are several families of genes whose divergence times differ depending on the taxonomical position of the animal (Klein 1986; Klein and Figueroa 1986; Kasahara et al. 1995). In primates, a few of the class I loci diverged prior to the emergence of this order, but most are of much more recent origin (Hughes and Nei 1989a). The primate class II gene families DO,DP, DQ, and DR, on the other hand, diverged before the radiation of the eutherian mammals (Carson and Trowsdale 1986). Within the families, the loci can vary considerably in their ages (Satta et al. 1996a,b). Loci of the DRB subfamily in particular appear to have undergone frequent expansions and contractions (Klein et al. 1993) that considerably obscured their evolutionary history. The deciphering of their history is further complicated by the fact that parts of the genes are subject to convergent evolution (Andersson et al. 1991; Kriener et al. 2000a,b). Interpreting the evolution of these genes is therefore a daunting task, which can succeed only if based on a combination of different approaches and utilization of a variety of marker systems.
In earlier publications (Kriener et al. 2000a,b), we provided evidence that exon 2 sequences, on which previous phylogenies of primateDRB genes were based (Trtková et al. 1993; Figueroa et al. 1994; Gyllensten et al. 1994), are providing misleading phylogenetic signals. The evolution of the exon is strongly affected by positive selection (Hughes and Nei 1989b), which creates repeatedly and independently similar sequence motifs (O'hUigin 1995; Kriener et al. 2000a,b). These motifs make genes appear more closely related than they are in reality. This, at least, is the message extracted from comparisons of the DRB intron with the DRB exon 2 sequences. Specifically, whereas the exon 2 sequences suggest that most primate DRB genes derive from a common ancestor that existed prior to the divergence of prosimians, Platyrrhini, and Catarrhini, intron sequences support the origin of DRB genes in each of the three taxa from a distinct ancestor (Kupfermann et al. 1999;Kriener et al. 2000a,b). Analysis of the exon 2 similarities implies molecular convergence as an explanation and thus indicates that the introns and not exon 2 are reflecting the true DRB gene phylogeny. However, as both the exon 2 and intron data are sequence–based, an independent source of information corroborating these conclusions was needed. We sought this source in the Aluelements inserted into the DRB genes.
The use of short interspersed repetitive elements (SINEs) in phylogenetic analysis is widespread. They have been used successfully to resolve phylogenies of a variety of mammals and other vertebrates (Batzer et al. 1994; Shimamura et al. 1997; Stoneking et al. 1997;Hamdi et al. 1999; Nikaido et al. 1999). They offer the advantages of ubiquity, uniqueness, and stability. Insertion occurs often enough to provide an array of useful cladistic markers. The chance of independent insertions at identical positions is small. Finally, SINEs are rarely removed without leaving evidence of their previous presence.
Alu repeats constitute one of several families of SINEs found in the mammalian genome (Deininger and Batzer 1993; Jurka 1995). They are believed to be derived from the 7SL RNA, which is a part of an 11S cytoplasmic ribonucleoprotein involved in targeting secretory proteins across the membranes of the endoplasmic reticulum (Ullu and Tschudi 1984). The derivation occurred in multiple steps in which two variants were first produced by deletions in different parts of the 7SL RNA gene — the free left Alu monomer or FLAM and the free rightAlu monomer or FRAM — and these monomers then fused to form the dimeric Alu elements (Quentin 1992a,b). All three forms are still found in the primate genome. The dimeric family ofAlu elements is divided into three major subfamilies that are of different age, and which, in the standardized nomenclature of Batzer et al. (1996), are designated J (∼80-million-years [my] old), S (∼35–44-my old), and Y (< 5-my old). Each subfamily is further divided into sub-subfamilies (e.g., the S subfamily is differentiated into Sx, Sy, Sp, and Sc branches). The subfamilies and the sub-subfamilies are distinguished by diagnostic substitutions shared by all members of a given group. The Alu elements are retroposons owing their mobility to the possession of sequences enabling their transcription by RNA polymerase III.
As Alu elements are ubiquitous in the primate genome, they can be identified relatively easily in any genomic region of interest. In earlier studies, we identified a series of > 60 Aluelements in the catarrhine Mhc–DRB genes (Schönbach and Klein 1991; Mňuková et al. 1994; Satta et al. 1996a) and designated them Alu1–Alu61. The aim of the present study was to identify platyrrhine and prosimian DRB gene-associatedAlu elements and use them to resolve the incongruences between the exon 2- and intron-based phylogenies.
RESULTS
In our search for Alu markers suited to the stated purpose, we focused on intron 1 because of its proximity to exon 2, which is the most variable of all the DRB exons, because of its length of several kilobase pairs (kb), which increases the likelihood of repeats' presence, and because several Alu elements were identified in it in catarrhine DRB genes (Andersson et al. 1987; Mňuková et al. 1994; Satta et al. 1996a), including one old element (Alu50J). To identify Alu elements in platyrrhine DRB genes, we selected seven genomic New World Monkeys (NWM) DNA samples bearing previously identified DRBexon 2 sequences (Trtková et al. 1993). Using exon 1- and exon 2-based primers, we then attempted to amplify the entire intron 1 and most of exon 2 of the different DRB genes by PCR. We succeeded in amplifying segments from 24 different DRB genes and failed with 5, perhaps because of a too large intron length. We confirmed the identity of the 24 amplification products by cloning them and sequencing their ends, including exon 2. For 23 of the 24 clones, the exon 2 sequences have already been described (Trtková et al. 1993; Gyllensten et al. 1994; Antunes et al. 1998; Kriener et al. 2000a, b), whereas one sequence identified a new exon 2, which we designate Sasc–DRB*W3401. Ten of the 24 amplification products were chosen for restriction digest and hybridization analysis.
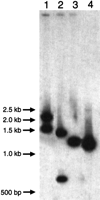
Samples of each of the 10 clones were divided into three parts and each part was digested with a different pair of restriction enzymes (BamHI–HindIII, HindIII–HincII, and BamHI–EcoRI). The digests were separated by gel electrophoresis, blotted, and the blots were hybridized with anAlu-specific probe (Fig. 1). The probe was obtained by PCR amplification of human genomic DNA using primersAlu1 and Alu2 (Table 1). It was∼ 250-bp long and was shown in control experiments to hybridize with members of the J, Sb, Sc, and Sq subfamilies ofAlu elements. By use of this probe, one or two hybridizing fragments could be identified on the blots of the digested NWM clones. The positive fragments were then subcloned and sequenced. The sequences were aligned and the Alu elements in them identified (Fig.2).
Southern blot analysis of a Saoe-DRB11*0102 intron 1 clone. The clone was digested with different pairs of restriction enzymes. The digests were separated by gel electrophoresis, blotted, and the blot hybridized with an Alu-specific probe. (Lanes 1–3) Digests obtained after treatment with theBamHI/HindIII, HindIII/HincII, andBamHI/EcoRI enzymes, respectively.(Lane 4) Positive control. A 1.2-kb fragment containing Alu50J was amplified from human genomic DNA and blotted.
Oligonucleotide Primers Used for PCR Amplification
Nucleotide sequence alignments of the Alu elements identified in the DRB genes of platyrrhini, strepsirrhini, and haplorrhini. The shaded boxes represent the flanking direct repeats that are created during the insertion of the Alu element. InAlu62 and Alu63, the diagnostic positions used for the subfamily classification are highlighted. A simple majority consensus sequence is given at the top. A dash (-) indicates identity with the consensus, an asterisk (*) an indel, and a dot (.) unavailability of sequence information. Numbering above the sequences starts with the first nucleotide of the alignment.
This approach revealed the presence of three distinct Aluelements in platyrrhine DRB intron 1 sequences. One of these three elements, Alu50, was identified previously in catarrhineDRB genes (Mňuková et al. 1994), the other two are new and so we designated them Alu62 and Alu63. The identification of the first element as Alu50 is based on sequence similarity and sharing of flanking direct repeats, as well as its position and orientation in intron 1 (Figs. 2 and3). By some of the same criteria, Alu62 andAlu63 are distinct from all other Alu elements identified thus far, which means that they are absent in all of the analyzed catarrhine DRB genes. The Alu50 element was found to be present in all 10 clones; Alu62 and Alu63were present in some of them and absent in others (Fig. 2).
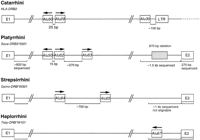
Diagram of intron 1 of selected catarrhine, platyrrhine, and prosimianDRB genes. The Alu elements identified in the intron are shown and their orientation is indicated by arrows. The distances between the Alu elements and the length of sequenced fragments in the 5‘ and the 3‘ ends of intron 1 are indicated by brackets. The shaded box represents a 870-bp deletion in the 3‘ end of intron 1. The drawing is not to scale.
The presence or absence of the three Alu elements among the remaining 14 of the 24 clones (i.e., those not subjected to restriction enzyme analysis) was established by PCR typing. To this end, the DNA isolated from the individual clones was PCR amplified by using different combinations of primers specific for each of the threeAlu elements (Table 1; Fig. 4). The specificity of the primers was based on the uniqueness of the sequences flanking the individual Alu elements. The typing identifiedAlu50 in all 14 clones; Alu62 in Saoe-DRB11*0105, Caja-DRB1*0307, Caja-DRB*W1602, Caja-DRB*W1603, Caja-DRB*W1605, Caja-DRB*W1612, Sasc-DRB*W1401, Sasc-DRB*W1901, Sasc-DRB*W3401, Ceap-DRB*W1301, and Ceap-DRB*W1502; andAlu63 in Saoe-DRB11*0105, Caja-DRB1*0307, Caja-DRB*W1602, Caja-DRB*W1603, Caja-DRB*W1605, Caja-DRB*W1612, Sasc-DRB*W1901, and Ceap-DRB*W3201. [The Alu50 element is present in all catarrhine DRB genes tested except Maar-DRB1*0301,in which the Alu50 region of intron 1 has been deleted (K. Kriener unpubl.). The Alu62 and Alu63 elements, as already mentioned, are absent in all identified catarrhine DRB genes.]
Location of the primers used for PCR amplifications. Primers (arrows) located in exon 1 and exon 2 were used to amplify the entire intron 1 of DRB genes. The cloned products were typed for the presence of Alu50, Alu62, and Alu63 with combinations of the primers Alu50-I–Alu50-VII and AluSc-I–AluSc-VI. The sequences of the primers are given in Table 1. (E) Exon, the shaded boxes represent Alu elements.
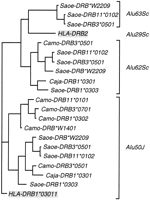
The Alu50, Alu62, and Alu63 elements are located in the same region of intron 1, arranged in this order in the 5' to 3' direction, ∼1.2 kb downstream from the 5' end of intron 1 (Fig. 3). Where all three elements are present on the same clone, Alu62 is immediately adjacent toAlu50 in a head-to-head orientation and Alu63 is ∼270 bp downstream of Alu62 in the same orientation asAlu62. In the same region, different Alu elements are found in both catarrhini (Alu29; Satta et al. 1996a) and prosimians (see below). The region therefore appears to be highly prone to Alu insertions. Sequence comparisons (Fig. 2) and analysis of diagnostic sites (Jurka and Milosavljevic 1991), as well as phylogenetic analysis of the sequences (Fig. 5), assign both Alu62 and Alu63 to the Sc subfamily.
Maximum likelihood tree based on the sequences of human and NWMAlu elements (Fig. 2). The classification of Alu62and Alu63 in the Sc subfamily is supported by their grouping with Alu29 of the HLA-DRB2 gene. Sequences ofAlu50 from the HLA-DRB1*03011 gene and ofAlu29 from the HLA-DRB2 gene are highlighted.
To investigate Alu elements of prosimian DRB genes, we used the exon 1- and exon 2-based primers on genomic DNA isolated from three prosimian species to amplify intron 1 of these genes by PCR. The amplification products, which ranged in length from 3.5 to 10 kb, depending on the species and the gene, were cloned, the clones digested with restriction enzymes, the fragments separated by electrophoresis and blotted, and the blots hybridized with an Alu–specific probe. The enzymes and the probe used were the same as those used in the study of the platyrrhine DRB genes. The weakly hybridizing fragments of five clones (three from Galago moholi and one each from Tarsius syrichta and T. bancanus) were subcloned and sequenced. The sequences revealed the presence of four different Alu elements. Three of them, found in theGalago, could be identified by sequence comparisons as dimeric type II repeats characteristic for galago genomes (Daniels and Deininger 1985, 1991). Because the three elements are distinct from each other and from all previously identified, DRB-associatedAlu repeats, we designate them Alu64, Alu65, and Alu66. Alu64 is located in a region corresponding to that occupied in the platyrrhini by Alu50, Alu62, and Alu63; Alu65 is found ∼700 bp downstream ofAlu64; Alu66 is located at the 3' end of intron 1. The Alu64 and Alu65 elements are present in the Gamo-DRB*W301 gene; the Alu66 element in the Gamo-DRB*W401 and *W501 genes. The fourth element, designated Alu67, was found in the two tarsier species; it resembled the recently described tarsier-specific type of elements (Zietkiewicz et al. 1999). It occurred at the 3' end of intron 1 in the Tasy-DRB*W101 andTaba-DRB*W201 genes. Neither Alu50, which is present in virtually all platyrrhine and catarrhine DRB genes, nor any of the other Alu elements identified previously inDRB genes could be found in any of the prosimian genes. The presence in the prosimian genes of a single copy of the sequence that flanks both ends of the Alu50 element (data not shown) indicates that Alu50 was apparently never present in these genes. The prosimian genes possess an entirely different set of Aluelements than the DRB genes in Platyrrhini and Catarrhini.
To determine the relationship of the Alu-element distribution to the phylogeny of the DRB genes, we superimposed the former on the latter in Figure 6. The tree in Figure 6 is based on ∼600 bp of sequence at the 5' end of intron 1 (Kriener et al. 2000a; Fig. 3), in addition to ∼500 bp of sequence flanking the Alu elements. The sequences of the Alu region were submitted to GenBank (accession nos. AF197226 – AF197240). The presence or absence of the Alu elements shows no correlation with either the species of origin of the DNA or the exon 2-based DRB gene classification as reflected in the gene designations. In contrast, the Alu element distribution correlates with the intron 1 phylogeny, which also correlates with the presence or absence of a previously described 870-bp deletion at the 3' end of the platyrrhine intron 1 (Kriener et al. 2000a,b; Fig. 3). Because the deletion is absent in all tested catarrhiniDRB genes, it apparently arose in an ancestral gene within the platyrrhine clade.
Comparison of a phylogeny obtained by sequence data with the distribution of cladistic markers. The neighbor-joining tree is based on the sequence of the 5‘end of intron 1 in addition to the sequence from the region surrounding the Alu elements. Major primate groupings (Catarrhini, Platyrrhini, Haplorrhini, and Strepsirrhini) correspond to clades indicated on the tree. The numbers at nodes indicate the percentage of recovery of that node in 500 bootstrap replications. The presence and absence of Aluelements and of an 870 bp-deletion at the 3‘end of intron 1 are indicated by + and − symbols, respectively. The names of unique Alu elements are given where they occur.
DISCUSSION
In the preceding text, we regarded the primate order as consisting of three monophyletic groups, the prosimians, the platyrrhines, and the catarrhines. Although the monophyly of the last two groups has never been seriously contested, that of the prosimians iscontentious (Martin 1990), and we used the traditional designation merely as a convenient way of referring to non-anthropoid primates. Recent molecular evidence, in fact, strongly bolsters the splitting of prosimians into Strepsirrhini and Haplorrhini, the latter being a sister group of Anthropoidea (Goodman et al. 1998). The Strepsirrhini include lemurs and galagos, the Haplorrhini the tarsiers. The existence of separate sets of Alu elements in galagos and tarsiers (Daniels and Deininger 1985, 1991; Zietkiewicz et al. 1999; the present report) provides additional evidence for this split. The apparent polyphyly of the prosimians must be taken into account when interpreting the results of the present study.
Considered in the context of previous work on primate DRBgenes in our laboratory (Trtková et al. 1993, 1995; Figueroa et al. 1994; Kriener et al. 2000a,b; Kupfermann et al. 1999) and in other laboratories (Slierendregt et al. 1992; Gyllensten et al. 1994; Knapp et al 1997; Antunes et al. 1998), the results described here lead us to two conclusions. The first conclusion is that in each of the four main primate groups (Strepsirrhini, Haplorrhini, Platyrrhini, and Catarrhini), there are multiple DRB loci present. This claim is best supported by data available for the Catarrhini, in which multiple loci have actually been assigned to their respective positions on genetic maps (Klein et al. 1991). In humans, for example, nineDRB loci are known to exist, although they never occur all together on one chromosome (Klein et al. 1991). For the Platyrrhini, Haplorrhini, and Strepsirrhini the claim is largely based on the detection of more than two DRB sequences per individual (Trtková et al. 1993; Figueroa et al. 1994; Gyllensten et al. 1994; Antunes et al. 1998; the present study). The organization of the loci is not known in any of these groups. The observation that different numbers of sequences are amplified from various individuals can, however, be taken as a hint that the number of loci per haplotype varies in the same manner as it does in human DRB haplotypes.
The second conclusion is that in each of the four primate groups, theDRB genes derive from a separate ancestral gene. The four ancestors all in turn derive from a single gene that was the last common ancestor of all primate DRB genes. The existence of an ancestral primate gene separate from the ancestral genes ofDRB loci in other orders of eutherian mammals is suggested first, by intron sequence-based phylogenies (Kriener et al. 2000a,b;Kupfermann et al. 1999; Fig. 6), and second, by the types and distribution of the DRB-associated Alu elements (Schönbach and Klein 1991; Mňuková et al. 1994; Satta et al. 1996a; the present study). The intron sequence data are fully congruent with the Alu distribution results. Each of the four primate groups has a monophyletic set of DRB intron sequences and each has a distinct set of Alu elements. The only shared element is Alu50 (a member of the oldest subfamily of elements), which is found in both the Platyrrhini and Catarrhini; all of the others are restricted in their distribution to only one of the four groups. The sharing of Alu50 supports the grouping of Platyrrhini and Catarrhini into Anthropoidea. The group restriction of the other Alu elements supports the monophyly of each of these groups and of the DRB genes in the Strepsirrhini, Haplorrhini, Platyrrhini, and Catarrhini. The group restriction is also found in the shorter downstream introns surveyed by Kupfermann et al. (1999). This distribution pattern is consistent with the assumption that new elements became inserted into the DRB genes in each of the four primate groups as they diverged from each of their common ancestors.
The above interpretation is seemingly contradicted by the exon 2-based phylogenies of the DRB genes (Trtková et al. 1993;Gyllensten et al. 1994; Figueroa et al. 1994). These phylogenies lead to the conclusion that separate allelic lineages at DRB loci diverged before the divergence of primates into the four taxonomical groups and have persisted to the present day. Responsible for the phylogenies are sequence motifs shared between exon 2 segments of not only primates, but often also different orders of eutherian mammals (Andersson et al. 1987; Gustafsson and Andersson 1994). However, as discussed elsewhere (Kriener et al. 2000a, b), there is now compelling evidence available in support of the notion that the sharing of motifs is the result of convergent evolution driven by positive selection on exon 2, which codes for the main functional part of the class II Mhc molecules. Exon 2 sequences are therefore not suitable for phylogenetic analyses of DRB genes. True phylogenies of these genes can be revealed by the intron sequences and corroborated by the distribution and identities of the Alu elements inserted into them.
METHODS
Source of DNA
Genomic NWM DNA was isolated from peripheral blood leukocytes of one cotton-top tamarin (Saguinus oedipus, Saoe; Universität Bielefeld, Germany), two common marmosets (Callithrix jacchus, Caja), one common squirrel-monkey (Saimiri sciureus, Sasc; both TNO Institute of Applied Radiobiology and Immunology, Rijswijk, The Netherlands), one black-capped capuchin (Cebus apella,Ceap), and two dusky titis (Callicebus moloch,Camo; both Universität Kassel, Germany). [The species abbreviations listed in parentheses are in accordance with the rules for standardized Mhc nomenclature (Klein et al. 1990)]. Prosimian DNA was prepared from one Philippine tarsier (Tarsius syrichta, Tasy), one Horsfield's tarsier (Tarsius bancanus, Taba; both CNRS Paris, France), and one moholi bushbaby (Galago moholi, Gamo; Universität Kassel). The DNA was extracted according to the protocol of Blin and Stafford (1976).
PCR
Fifty to one hundred nanograms of genomic DNA were amplified with 0.5 μM of each of the two primers (Table 1; Fig. 4), 200 μM of each of the four deoxyribonucleotide phosphates, and 1.5 mM MgCl2in the form of Hot Wax Mg2+ beads (Invitrogen, Leek, The Netherlands) using the GeneAmp XL PCR Kit (Perkin Elmer Applied Biosystems, Foster City, CA). The amplification was performed in the Gene Amp PCR System 9600 (Perkin Elmer Cetus, Norwalk, CN) and consisted of 12 cycles of denaturation at 94°C for 30 sec, followed by annealing and extension at 64°C for 8 min, then 24 cycles, in which the annealing temperature was raised by 0.15°C in every cycle. The reaction was completed by a final primer extension for 10 min at 72°C. The amplification products were purified and cloned in theSmaI site of the pGEM-3Zf(+) (Promega, Madison, WI) or the pUC18 plasmid vector (Amersham Pharmacia Biotech, Freiburg, Germany).
DNA Sequencing
Double-stranded DNA was prepared with the QIAgen Plasmid Kit (Qiagen, Hilden, Germany) and sequenced by using the AutoRead Sequencing Kit (Amersham Pharmacia Biotech). Five microliters of each sequencing reaction mixture were loaded on a 6.6% acrylamide gel and run in the Automated Laser Fluorescent DNA sequencer (Amersham Pharmacia Biotech). Cycle sequencing reactions were performed with the 7-deaza-dGTP Kit (Amersham Pharmacia Biotech) and run in the LiCor sequencer (MWG Biotech, Ebersberg, Germany).
Restriction Enzyme Digestion and Southern Blotting
Clones were digested with the restriction enzyme combinationsBamHI/HindIII, HindIII/HincII, andBamHI/EcoRI (NEB, Beverly, MA; Boehringer, Mannheim, Germany) according to the manufacturer's instructions. After 1 hr of incubation, the digestion products were separated on a 1% agarose gel and transferred to positively charged nylon membranes (Hybond N+, Amersham Pharmacia Biotech or Gene Screen Plus, NEN, Boston, MA) by alkaline vacuum blotting with 0.4 N NaOH as a transfer solution.
Hybridization
A hybridization probe was obtained by PCR amplification of human genomic DNA using primers Alu1 and Alu2. The primers bound to the 5‘ and 3‘ ends of a dimeric Alu element, respectively, and amplified a 250-bp fragment. The PCR product was labeled with α[32PdCTP] by using Ready-To-Go DNA labeling beads (Amersham Pharmacia Biotech). Prehybridization was carried out for 1 or 2 hr at 42°C in a solution containing 50% formamide, 5x SSPE, 5x Denhardt’s solution, 0.1% SDS, and 100 μg/ml sonicated salmon sperm DNA. The hybridization probe was denatured and added to a fresh hybridization solution. Hybridization was carried out overnight at 42°C. The membranes were washed twice for 15 min at room temperature in a solution containing 2x SSPE and 0.2% SDS, and once for 15 min at 50°C in a solution containing 0.5x SSPE and 0.2% SDS. The filters were then used to expose X-ray film (XAR5; Kodak, Stuttgart, Germany). The hybridization-positive restriction fragments thus identified were subcloned and sequenced.
Sequence Analysis and Classification of Alu Elements
Sequences were scanned by using the program Dotty Plot, version 1.0c (Gilbert 1995a) and aligned with the help of the program SeqPup version 0.4 (Gilbert 1995b). Genetic distances were calculated by the two-parameter method (Kimura 1980). Phylogenetic trees were drawn by the neighbor-joining method (Saitou and Nei 1987) in the version specified by the program MEGA (Kumar et al. 1993) and by the maximum likelihood method using the program PHYLIP (Felsenstein 1993).Alu elements were aligned to the consensus sequence ofAlu elements and their flanking direct repeats were identified. They were classified in subfamilies according to Jurka and Milosavljevic (1991).
Acknowledgments
We thank Ms. Jane Kraushaar for editorial assistance, Dr. Herbert Tichy, MPI für Biologie, Tübingen for the NWM and prosimian DNA samples, and Dr. Philippe Dijan, CNRS, Paris for providing us with tarsier DNA. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL jan.klein{at}tuebingen.mpg.de; FAX 49 7071/600437.
-
- Received October 4, 1999.
- Accepted January 18, 2000.
- Cold Spring Harbor Laboratory Press