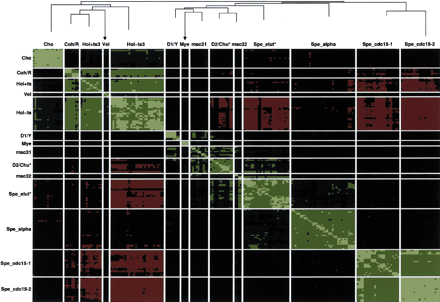
Results of clustering 217 conditions by Pearson correlation coefficients over 1078 ORFs, plus high-level dendrogram showing the 14 highest level condition clusters and their relationships in the clustering hierarchy. The 1078 ORFs exhibited high median relative abundance over all conditions and showed evidence of induction or repression (see Methods). Conditions are presented symmetrically as lines and columns of cells with each cell representing the correlation coefficient between two conditions over the log10 relative abundances of the ORFs, expressed in standard units for the ORF over all conditions. Red values indicate negative correlation coefficients and green values indicate positive correlation coefficients. Brighter red (green) values indicated more negative (positive) correlation. Diagonal entries all represent correlation coefficients = 1 and are bright green. Dendrogram branch heights from top of the tree indicate relative locations of the join creating the subcluster in the sequence of subcluster agglomerations that created the tree; thus, clusters at the end of longer branches may be considered more similar than clusters at the end of shorter branches from the perspective of the clustering algorithm. Arrows indicate branches that had to be truncated for this diagram. Cluster symbols are series codes (see Table 1) except for the following: Coh/R = Coh + Rot (R). Hol+ts3 = all Hol conditions involving temperature-sensitive mutants except for Hol_med6_ts_1, which is in the Hol∼ts3 group. (A replicate condition Hol_med6_ts_2, however, is in Hol+ts.) Hol∼ts3 = all Hol conditions that do not involve temperature-sensitive mutants excepts for Hol_med6_ts_1. This cluster contains all control series as well as all non-temperature-sensitive mutants. Other labels are for microarray-derived data sets. These are discussed on our web site.











