Systematic Management and Analysis of Yeast Gene Expression Data
Abstract
We report steps toward the systematic management, standardization, and analysis of functional genomics data. We developed the ExpressDB database for yeast RNA expression data and loaded it with ∼17.5 million pieces of data reported by 11 studies with three different kinds of high-throughput RNA assays. A web-based tool supports queries across the data from these studies. We examined comparability of data by converting data from 9 studies (217 conditions) into mRNA relative abundance estimates (ERAs) and by clustering of conditions by ERAs. We report on generation of ERAs and condition clustering for non-microarray data (5 studies, 63 conditions) and describe initial attempts to generate microarray-based ERAs (4 studies, 154 conditions), which exhibit increased error, on our web sitehttp://arep.med.harvard.edu/ExpressDB. We recommend standards for data reporting, suggest research into improving comparability of microarray data through quantifying and standardizing control condition RNA populations, and also suggest research into the calibration of different RNA assays. We introduce a model for a database that integrates different kinds of functional genomics data, Biomolecule Interaction, Growth and Expression Database (BIGED).
Ever-growing amounts of sequence data for numerous organisms, combined with readily available technology for large-scale expression studies on the basis of oligonucleotide arrays, DNA microarrays, Serial Analysis of Gene Expression (SAGE), and other techniques (Velculescu et al. 1995; Lockhart et al. 1996; DeRisi et al. 1997), has led to the rapid accumulation of large expression data sets and the development of the field of functional genomics. Functional genomics has been contrasted as having a systematic, genome-wide approach to the collection and analysis of biological data compared with more traditional methods, which focus in depth on particular genes, proteins, or pathways (Hieter and Boguski 1997). But despite rapid strides, capped by a string of successful studies (see Table1), functional genomics has yet to develop a highly integrated system of tools and methods such those used in sequence analysis and structural genomics (Hieter and Boguski 1997); for that, we must await development of the three following components: general databases, data standards, and integrated general-purpose analysis tools. A comparison of the status of these fields with respect to these components is given in Table 2. Note that throughout this article sets of related experiments and conditions assayed within them will be denoted by series codes, which are defined in Table 1.
Yeast RNA Expression Data Sets Incorporated into Data Analysis
Comparison of Status of Functional Genomics
As these three components are dependent on each other and must coevolve, the right mixture will only come together with experience. We hope to jump start the process by describing here the working prototypes of parts of the required systems and examples of what can be done with them. Specifically, we describe ExpressDB, a general database for RNA expression data that has been loaded with data from 11 yeast studies using three different kinds of high-throughput RNA level assays (see Table 1). We also describe EXD, an integrated web-based application that supports user queries of ExpressDB data. ExpressDB and EXD differ from existing research-specific databases (see web sites on Table 1) in that they represent and manage data from multiple studies, and complement databases such as ArrayDB (Ermolaeva et al. 1998) by managing data from multiple kinds of RNA level assays.
In each study whose data were loaded into ExpressDB, data were collected and prepared in ways appropriate to the study's particular experimental design. Because designs and methods are not generally coordinated across studies, data from different studies are not always easily compared. This has no impact on the success of each study individually, but data comparability assumes increased importance in a database context in which comparability improvements can translate into simpler and more meaningful queries, more efficient database structures, and opportunities for more effective data mining. To gauge comparability of currently available data, we therefore formulated what we considered to be an attainable ideal and assessed what would be required for the data on ExpressDB to meet it. We propose specific recommendations on the basis of this assessment (see Discussion). We defined our ideal state of data comparability to be: (1) All RNA expression data are provided in the form of estimated relative abundances (ERAs) of a defined set of functionally distinguishable RNA fragments (FDRs) which, in the present case, we take to be RNAs corresponding to ORFs. An ORF ERA represents the fractional abundance of the ORF's RNA with respect to the total population of ORF RNAs in cells in a particular experimental condition (defined by cell strain and environmental history). (2) Analytical tools used to measure data comparability confirm that similar conditions have similar RNA expression profiles, regardless of which RNA assays are used for data collection. The rationale for (1) is that ERAs are intuitive and unambiguous measures of RNA level that are theoretically directly comparable across conditions regardless of experimental methodologies.
In assessing ExpressDB data against this ideal, we converted as much data on ExpressDB as possible to the form of ERAs, and explored clustering of conditions by ORF expression profiles as a tool for analytically investigating data comparability. We undertook these steps for data generated from oligonucleotide arrays (4 studies, 60 conditions) SAGE (1 study, 3 conditions), and microarrays (4 studies, 154 conditions), generating a set of ORF ERAs for 217 conditions. Issues with microarray data currently make it difficult to compute ERAs from them and our best effort resulted in microarray ERA values that exhibit increased variability compared with corresponding ratios (coefficient of variation of ERAs = 3.3 times that of ratios) (see Results). We focus here on methods and results for Affymetrix and SAGE data. Readers interested in our microarray results may consult the supplemental material on our web sitehttp://arep.med.harvard.edu/ExpressDB. A file containing ERAs for 213 conditions—all except for four not previously published (Coh)–may be downloaded from the same web site. The ERA file has not been loaded into ExpressDB, which contains only original data, but has been loaded into a separate database on our web site to allow it to be queried easily.
RESULTS
Database
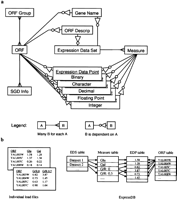
ExpressDB is a relational database for RNA expression data. We implemented it using Sybase SQL Server 11.0.3 on a shared DEC 3000 server running DEC Unix 4.0D. We conceive of ExpressDB as a generalized two-dimensional table that can subsume individual tables of expression data reported by researchers. We provide a high-level logical data model for the ExpressDB database and an example of how it operates as a generalized two-dimensional table in Figure 1. That figure also presents names of ExpressDB tables that will be used throughout this article. Note that names of these tables are always capitalized (e.g., Measure).
Logical data model of ExpressDB and its usage as a generalized two-dimensional table. (a) High level logical data model. Rectangles represent tables in the database and connecting lines represent database table relationships as described in the legend and as per Teorey (1994). (b) Use of Express DB as a generalized two-dimensional table. Database load files are usually in the form of tables with each line dedicated to an ORF or control probe and each column to a measurement, computed value, or descriptive field. The ExpressDB ORF table contains records for all ORFs or control used by any load file, and each measurement column from each load file is represented by a Measure record. Measure records associated with a related set of experiments, especially measures reported by a single literature reference, are all linked via a database relationship to a common Expression Data Set record, although multiple Expression Data Set records are used for some references. Measurement column values for each ORF for an experimental data series are given in Expression Data Point records that are linked to their ORF and Measure records via database relationships. To accommodate different formats of column values, five different Expression Data Point tables are available, each supporting a different format.
We developed a utility program EDBUpdate to load data from individual tab-delimited files (load files) into ExpressDB. Load files must present a systematically collected set of measurements or descriptive information for a series of ORFs, in which each line of the file presents information for an ORF and each column a particular measurement or information field. Examples of measurements are numerical values representing ORF mRNA abundance and data-quality indicators. An ORF description field would be an example of an information field. Measurement columns are represented by ExpressDB Measure records. Using EDBUpdate, we performed loads of all available data files associated with the studies described in Table 1 into the database as well as data from two others (Eisen et al. 1998; Marton et al. 1998). ExpressDB contains >17.5 million pieces of information. Other statistics on the database are given in Table 3.
ExpressDB Statistics
Load files downloaded from public sources often required minor editing to put them into the proper format for loading. For instance, files presenting data collected with Affymetrix oligonucleotide arrays often give the ORF and common gene name in the same column separated by a (/), and we had to separate these into distinct columns. More extensive work was required to load the SAGE-based expression data from Vel because data from SAGE is in the form of counts of tag sequences in cDNAs, whereas ExpressDB imposes as a structural requirement that data be indexed by ORF. A key issue for this indexing is that some SAGE tag sequences cannot be assigned to a unique ORF. As a result, for each SAGE condition, we computed and loaded into ExpressDB both a minimum and a maximum tag count for each ORF, in which the minimum count includes only counts of tags uniquely assignable to the ORF (which we call unambiguous tags) and the maximum count includes these plus the counts of tags shared with other ORFs (which we call ambiguous tags). Additional details may be found on the ExpressDB database in the Expression Data Set record for the Vel experiments.
Database Query Application
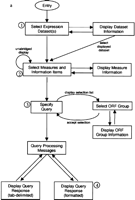
Our web-based query interface for ExpressDB, the EXD system, can be accessed at http://arep.med.harvard.edu/ExpressDB/. A JavaScript 2.1-supporting web browser such as Microsoft Internet Explorer 4.0+ or Netscape Navigator 4.0+ is required. The logical flow of the EXD system is depicted in Figure 2a. The main line of this logic is that the user is prompted for successively more detailed specifications concerning the query, starting with the Expression Data Sets (see Fig. 1) of interest, moving on to the Measures of interest within these Expression Data Sets, and finally to conditions that must be satisfied by the ORFs or their Measure values. ExpressDB allows Expression Data Set and Measure records to be marked private and these are not offered for user selection by EXD; this option has been used for the Coh set of experiments that are not yet published. The query conditions offered for user specification are sensitive to the data format of the Measures; thus the user is prompted for text matching specifications when a Measure has a character format, and with numerical equalities and inequalities for numerical formats. Statistical specifications may also be indicated for numerical measures, for example, it is possible to ask for all ORFs for which the value of a measurement is greater than two standard deviations from the mean. It is also possible to ask for only those ORFs that are either in or not in a group of ORFs defined in the ExpressDB ORF Group table. To demonstrate this capability, we loaded this table with 207 functional groupings of yeast ORFs defined on the Munich Information Center for Protein Sciences (MIPS) database (Mewes et al. 1999). The output of ExpressDB is given in either a tab-delimited or formatted form. Tab-delimited output can be copied from the screen and pasted into desktop applications like Excel (Microsoft, Redmond, WA) for further analysis. We consider this a rudimentary form of integration with and pipelining to downstream computer analysis tools. Additional information on querying the database can be found on the web site mentioned above.
Logical flow through the EXD query system and example of EXD query. (a) Logical flow through EXD query system. Boxes correspond to forms and web pages produced by the system. Arrows correspond to movements through the system available through hotlinks and buttons on the forms and pages. Number in circles indicated forms and web pages depicted in the example of an EXD query in b of this figure. (b) Illustration of an EXD query provided to give an impression of EXD system usage. (1) A user selects two data sets from the database, (2) selects an information field (SGDID) and nine Measures from these data sets, (3) specifies a query condition, and (4) obtains results. Forms and web pages shown in diagram are versions of actual EXD pages that have been reduced in size, abridged, and edited to highlight key features. Details pertaining to the example are given in the text.
Figure 2b provides an impression of what it is like to use EXD to perform a typical ExpressDB query. On entry to the system, the user is presented with a form listing data sets available on the database (Fig.2b, step 1). The user selects one or more data sets; here the Der_diaux and Der_tup data sets have been selected (see Table 1). On clicking the Submit button, the user is brought to the next form (Fig. 2b, step 2) which presents information fields and Measures available on the database for the selected data sets, and the user chooses the ones he or she wishes to see. Here, the user has asked to see the information field SGDID (Saccharomyces Genome Database identifier) (Cherry et al. 1999) and all of the ratios from the two selected data sets (two from Der_tup and seven from Der_diaux) that represent an ORF's fold change of mRNA abundance in an experimental condition relative to its control condition. On clicking the Submit button, the user is next brought to a form (Fig. 2b, step 3), which allows entry of query specifications. In this example, the only specification provided is that the microarray ratio from the last Der_diaux condition must be >1 s.d. above the mean for this Measure. This will cause EXD to display all selected information fields and Measures for only those ORFs meeting this specification. The biological meaning of this particular specification is that the user wishes to see data for only those ORFs that are at least moderately induced in ethanol, as the last Der_diaux condition represents the end of a diauxic shift time series during which the yeast cells have consumed all of the glucose originally available in the medium and are growing on ethanol at the end of the shift (DeRisi et al. 1997). This illustrates the level of knowledge that a user must have of the meaning of the data sets and Measures on the database to make effective use of the EXD system. The design of the database allows descriptive information about data sets and Measures to be stored so that users may obtain this knowledge directly from the database itself. When the user clicks Submit on the query specification form, the EXD system processes the query. This may take several minutes, during which processing messages are displayed by the system (not illustrated in Fig. 2b). When processing is complete, the user clicks on a hotlink to see the results (Fig. 2b, step 4).
We believe the EXD system to be the first query system that allows users to query simultaneously any of the expression data reported by experiments associated with different literature references and return the results collated by ORF name. Fundamentally, this derives from the fact that all of the data has been collected in one database, but it is also supported by EXD's ability to navigate ExpressDB's generalized two-dimensional table structure. At this time, however, we recommend use of EXD only for relatively simple queries involving ∼10 or fewer Measures over all or a group of ORFs, partly because of performance issues with more complex queries and the database's shared computer environment, and partly because we need to develop an interface that makes it easier for users to find data items of interest from a set of >2000 available Measures and then specify query conditions for them.
Generation of ERAs
Generation of ERAs is straightforward for data derived from Affymetrix oligonucleotide arrays and SAGE (see Methods), but microarray-derived data present a significant issue. Microarray-based experiments simultaneously collect intensity levels of fluorescently labeled cDNAs derived from an experimental condition, and intensity levels of cDNAs, labeled with a different fluorophore, derived from a control condition. The two cDNA preparations are hybridized in parallel to the same probe sequence spots on the array (DeRisi et al. 1997). Ratios between background-subtracted experimental and control condition intensities are used for data reporting and analysis because they compensate for several sources of bias and noise in intensity results, including ORF-to-ORF variations in labeled nucleotide incorporation (bias), ORF-to-ORF variations in efficiency of the PCR reactions used to generate the probe sequences spotted onto the arrays (bias and noise), and spot size and shape variations (noise). Microarray ratios differ from ERAs in that they are fold changes of an ORF RNA level in an experimental condition relative to a control condition, whereas ERAs are fractional abundances of an RNA in a single condition. Moreover, microarray ratios cannot be converted into ERAs; when total RNA levels are roughly constant, microarray ratios may be identified with ratios of experimental condition ERAs to control condition ERAs, neither of which can be determined from the ratios without prior knowledge of the other. Affymetrix- and SAGE-based experiments are not subject to this difficulty because neither require measurements relative to a control condition. Experiments using Affymetrix arrays typically report ORF expression level measurements as averages of differences in background-subtracted intensities of fluorescently labeled sample cRNA or cDNA hybridized to probe sets of ∼20 perfect match (PM) and mismatch (MM) oligonucleotide probes for the ORF (average PM–MM). Sequence rules for probes and the precision of the oligonucleotide synthesis process control for noise and bias, reducing the need for reporting relative to a control condition RNA sample; mismatch probes, which differ from their perfect match counterparts by a single base, control for cross-hybridization (Lockhart et al. 1996). SAGE experiments rely on sequencing of cDNA tag sequences rather than hybridization, eliminating probe-, hybridization-, and label-based variation, and use procedures that control for tag sequence amplification biases (Velculescu et al. 1995, 1997). Again, only a single condition's RNA sample is required. As noted above, although we computed ERAs for data sets derived using all three RNA assays, we only report here on generation of ERAs for Affymetrix- and SAGE-derived data. A discussion of microarray-derived results may be found on our web site.
Other issues that complicate both generation of ERAs and comparisons of data generally include (1) the frequent reporting (see Table 3) of multiple measurement values for an ORF from single experimental or control conditions derived from multiple spots for an ORF on a microarray or multiple probe sets for an ORF on an Affymetrix array, which raises the question of how values should be combined or selected for further analysis, and (2) use of different ORF names across different sets of experiments, making matching of ORF data across experiments difficult.
In Affymetrix-based experiments, multiple values for an ORF arise from distinct probe sets for the ORF that are distinguished by their probes being located to different exons or other general probe set characteristics. We found different types of probe sets to have different properties. For instance, we computed an aggregated measure (see Methods) of the ratio of average PM–MM values of exon 1 probe sets against those of exon 2 probe sets from values given in the Hol and Cho sets of experiments, and found that in both cases, exon 1 probe set values were, in aggregate, ∼0.6 of exon 2 probe set values (Hol:N = 90 ratios = 96 ratios–6 outliers; Cho:N = 93 ratios = 100 ratios–7 outliers;s.d. = 0.4 for each distribution). When presented with a choice of expression measurements for the same ORF with different values, one would ideally like to identify and use the measure of highest quality, but the fact that exon 1 probe sets have the property of yielding smaller measurement values than exon 2 probe sets does not imply that exon 1 probe sets are of less quality. Our strategy for consolidating multiple Affymetrix probe set values therefore focused on consistency. Because most probes for ORFs with single probe sets are taken from the 3′ ends of ORF sequences, we decided to handle ORFs with probe sets for multiple exons by using the exon 2 values instead of exon 1 values. We also avoided probe sets with special feature set indicators where possible (see Methods). Affymetrix GeneChip software returns a “presence call” that describes when a gene product may be considered to be present, marginally present, or absent in an RNA sample (Lockhart et al. 1996). An alternative strategy would have been to consolidate multiple probe set values by taking averages of all probe set values called as present for an ORF. However, we found exon 1 probe sets to be called absent significantly more often than exon 2 probe sets: The average of number of absence calls, ± thes.d. of the average, for an exon 1 probe set over 42 Hol conditions = 7.6 ± 1.1 versus 4.4 ± 1.0 for exon 2 probe sets. Our exon-based consolidation strategy therefore already, at least partially, takes presence calls into account while avoiding complications that would arise with presence call-based strategy: (1) Not all Affymetrix-based experiments report presence calls, (2) it often happens that multiple probe sets for an ORF are all marked absent whenever any one of them is (40% of 1594 multiple probe set values across the 42 Hol experiments).
In the case of SAGE, multiple measurements for an ORF arise from counts for distinct ORF SAGE tags. The sum of counts for unambiguous tags for an ORF, maintained as minimum tag counts on ExpressDB, can be safely attributed to RNA expression by that ORF, but counts for ambiguous tags included in maximum tag counts cannot be safely attributed to that ORF, as they may have come from the RNAs of different ORFs that happen to share the same tag (Velculescu et al. 1997). To assure the most accurate possible ERAs for SAGE conditions, we therefore only computed them for ORFs, all of whose tags were unambiguous. The number of ORFs for which we computed ERAs in the three Vel SAGE conditions (2122, 2211, and 2218) is thus considerably smaller than the number we computed for other conditions (5803 ± 585 for all 217 conditions). The ultimate origin of tag ambiguity in SAGE, sequence similarity between genes, also affects oligonucleotide array and microarray measurement of gene expression through cross-hybridization of cRNAs and cDNAs to ORF probe sets or spots.
In the end, we produced a file containing ERAs for all ORFs for which there were usable data for 217 conditions (60 Affymetrix, 3 SAGE, and 154 microarray). The number of ORFs (identified by name) for which data are provided in at least one condition is 6293. The process of generating ERAs included steps to resolve different names for the same ORF (see Methods), and of these 6293 all but 94 could be identified with SGDIDs . Wherever data for an ORF was not reported in a condition, or an ERA could not be computed for an ORF, a “null” value (empty field) is included in the table for that ORF and condition. Because the maximum number of ORFs for which ERAs are reported in a condition is 6221, all conditions contain some null values; some conditions, like the SAGE conditions noted above, contain large numbers of null values. As noted above, the version of the file that may be downloaded from our web site contains only 213 conditions (56 Affymetrix) because four conditions (Coh) have not been published previously.
Clustering of Experimental Conditions
Although clustering of ORFs on the basis of expression levels over sets of conditions has often been reported (Cho et al. 1998; Eisen et al. 1998; Wen et al. 1998; Tavazoie et al. 1999), clustering of conditions is less common but of increasing interest, in part because of its potential for classifying tumors (Weinstein et al. 1997; Alon et al. 1999; Perou et al. 1999). Here we used condition clustering to investigate its potential as a measure of comparability of data. The motivation for considering condition clustering in this role is that cells of similar strains in similar environmental conditions should exhibit similar ORF RNA abundances, and therefore similar conditions should yield high correlation coefficients and small Euclidean distances between their ORF abundance profiles. Condition clustering provides a convenient way of seeing such relationships in correlations and distances for large numbers of conditions. We therefore hypothesized that we should be able to find instances in which clearly similar conditions were clustered together and clearly dissimilar conditions were separated into different clusters. We also used condition clustering to assess the preliminary ERA values we generated for microarray conditions and to compare them against microarray ratios. Clustering of microarray ERA data is affected by the increased variability of these preliminary values. However, we found indications that condition clustering of microarray ratio data may be subject to biases when clustering conditions from sets of experiments using different control conditions. These biases did not appear in the clustering of corresponding microarray ERAs. We discuss these results on our web site.
When clustering ERA data, we should generally expect that conditions will tend to segregate into clusters according to related series of experiments for two reasons: First, conditions in related series frequently use the same or similar strains and cell environments. Second, differences in technique and equipment used in different studies may have the effect of weighting individual ORF abundances from different series differently. Both of these factors will tend to make conditions in a related series more similar in ORF ERA profile than conditions from different series. A diagram depicting the highest level 14 clusters of 217 conditions grouped by similarity of pairwise correlation coefficients over transformed ORF estimated relative abundances (see Methods) is shown in Figure 3. It is evident that conditions cluster mainly with other conditions in the same related sets of experiments. To confirm that this and other observations below are not simply artifacts of the clustering algorithm, we also performed clustering by an alternative method, the clustering of conditions directly by transformed ORF ERAs rather than pairwise correlation coefficients of conditions over their ERAs (see Fig. 4 in the supplemental materials on our web site). In both exercises, we clustered subsets of high-expressing ORFs that showed evidence of induction across conditions, rather than clustering over all ORFs, to reduce noise that might be introduced from large numbers of low-expressing ORFs (see Methods). Despite some shuffling of clusters at the highest levels, it remains true that conditions in the same related sets of experiments are found to be closer to each other than to conditions in other sets. Details may be found on our web site.
Results of clustering 217 conditions by Pearson correlation coefficients over 1078 ORFs, plus high-level dendrogram showing the 14 highest level condition clusters and their relationships in the clustering hierarchy. The 1078 ORFs exhibited high median relative abundance over all conditions and showed evidence of induction or repression (see Methods). Conditions are presented symmetrically as lines and columns of cells with each cell representing the correlation coefficient between two conditions over the log10 relative abundances of the ORFs, expressed in standard units for the ORF over all conditions. Red values indicate negative correlation coefficients and green values indicate positive correlation coefficients. Brighter red (green) values indicated more negative (positive) correlation. Diagonal entries all represent correlation coefficients = 1 and are bright green. Dendrogram branch heights from top of the tree indicate relative locations of the join creating the subcluster in the sequence of subcluster agglomerations that created the tree; thus, clusters at the end of longer branches may be considered more similar than clusters at the end of shorter branches from the perspective of the clustering algorithm. Arrows indicate branches that had to be truncated for this diagram. Cluster symbols are series codes (see Table 1) except for the following: Coh/R = Coh + Rot (R). Hol+ts3 = all Hol conditions involving temperature-sensitive mutants except for Hol_med6_ts_1, which is in the Hol∼ts3 group. (A replicate condition Hol_med6_ts_2, however, is in Hol+ts.) Hol∼ts3 = all Hol conditions that do not involve temperature-sensitive mutants excepts for Hol_med6_ts_1. This cluster contains all control series as well as all non-temperature-sensitive mutants. Other labels are for microarray-derived data sets. These are discussed on our web site.
To assess the ability of condition clustering to capture similarities and differences between experiments, we examined the
Hol set of 42 conditions. This set comprises 21 experiments with RNA polymerase complex mutants and 21 corresponding wild-type
controls. Within this set of experiments, (1) the 21 experiments contain 10 pairs of replicated experiments, and likewise
the 21 controls contain 10 pairs of replicates, making a total of 20 pairs of replicated conditions. Nine of these 20 pairs
of replicated conditions are clustered at the leaf level in Figure 3(P =
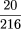 ×
×
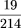 × · · · ×
× · · · ×
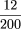 = 8.4 × 10−11), and 12 of these 20 are clustered at the leaf level in that depicted in Figure 4 (P =
= 8.4 × 10−11), and 12 of these 20 are clustered at the leaf level in that depicted in Figure 4 (P =
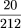 ×
×
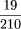 × · · · ×
× · · · ×
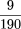 = 1.4 × 10−14). The significance of two conditions clustering at the leaf level is that they are more similar to each other than to any
other conditions. (2) The Hol series contains 13 conditions involving temperature-sensitive RNA polymerase complex mutants
that were maintained at 37°C prior to RNA assay. All other mutants and all control conditions were maintained at 30°C (see
Hol web site listed in Table 1). In the clustering of Figure 3, 12 of the temperature-sensitive mutant conditions segregate into their own cluster apart from the rest of the Hol series.
In Figure 4 (on our web site), all 13 temperature-sensitive mutant conditions segregate into a cluster of 15 that also contains
two control conditions. These clusters evidently reflect temperature effects. Together, these observations indicate that condition
clustering is effective at identifying similar (here replicated) conditions, and that it is likewise effective at segregating
conditions on the basis of important environmental variables such as temperature.
= 1.4 × 10−14). The significance of two conditions clustering at the leaf level is that they are more similar to each other than to any
other conditions. (2) The Hol series contains 13 conditions involving temperature-sensitive RNA polymerase complex mutants
that were maintained at 37°C prior to RNA assay. All other mutants and all control conditions were maintained at 30°C (see
Hol web site listed in Table 1). In the clustering of Figure 3, 12 of the temperature-sensitive mutant conditions segregate into their own cluster apart from the rest of the Hol series.
In Figure 4 (on our web site), all 13 temperature-sensitive mutant conditions segregate into a cluster of 15 that also contains
two control conditions. These clusters evidently reflect temperature effects. Together, these observations indicate that condition
clustering is effective at identifying similar (here replicated) conditions, and that it is likewise effective at segregating
conditions on the basis of important environmental variables such as temperature.
DISCUSSION
The database and query tool described here represent preliminary versions of tools required in an integrated tool kit for exploring expression data. They can be modified to make them more sophisticated and complete. Some improvements involve relatively simple technical fixes. The current version of ExpressDB is yeast specific, but the design changes required to generalize it are small and an organism-general version will soon be available. The key changes allow results to be recorded for FDRs other than ORF RNAs, such as ESTs, cDNAs, and noncoding RNAs, that are frequently reported for higher organisms, and allow different sets of FDRs to be registered for different organisms. The EXD query system can also be modified to automatically pipeline results to downstream analysis tools such as clustering by gene and condition. Other technical issues, such as system performance, will require ongoing management. Whereas database software and application tuning and equipment upgrades can improve ExpressDB's current performance, its current 17.5 million records, resulting from only 11 sources, clearly only represent the tiny beginnings of an anticipated flood of expression data. Over time, more efficient database technologies and algorithms will need to be explored to ensure maintenance of performance levels. Standardization of data formats and contents (see below) will also help improve performance by providing opportunities to structure the database more efficiently.
More involved issues raised by data comparability concerns must be addressed through standards and additional research. Here we propose several directions for development on the basis of our results. Because these directions apply beyond of the case of yeast, we phrase them in terms of FDRs rather than ORFs
Develop Methods that Will Allow Sets of Microarray-Derived Expression Data to Be Directly Compared with Each Other and with Sets of Expression Data Obtained Using Other Methodologies
By dint of its flexibility, relatively low cost, and public availability, microarray technology has made a huge contribution to both the science of functional genomics as a whole and to the number of RNA expression data sets available for analysis; but the full potential of these data will not be realized until methods are developed that allow microarray-derived ratios of FDR levels in experimental conditions relative to control conditions to be easily and directly compared with microarray-derived ratios on the basis of different control conditions, and with the results of other high-throughput RNA assays. One possibility would be to encourage the development of standard microarray control conditions. If the RNA species in such standards are quantified for abundance, it would then also be possible to generate ERAs from microarray-derived data. Some ideas for this are discussed on our web site.
Test Different RNA Expression Assays on Common RNA Samples to Determine Whether They Produce Equivalent Results, and Develop Standard Calibrations Where They Do Not
It is not enough that expression data collected with different methodologies be expressible in a common form such as ERAs; the actual data values must be shown to be equivalent regardless of their methodology of origin. We foresee a research project in which RNA extracts from several test combinations of strains and conditions are assayed on all key RNA expression assays. Condition clustering of results by test sample regardless of assay may be one good indicator of comparability of results and of the correctness of calibrations. Protocols for sample preparation and labeling may also need to be considered, as these, too, may influence comparability of results. For instance, among Affymetrix-based experiments, Cho generated labeled double-stranded cDNAs, whereas Hol, Rot, and Coh generated labeled single-stranded cRNAs. With the cDNA protocol, cDNAs from nearby adjacent or overlapping ORFs, especially convergent ORFs with 3′-end overlaps, could hybridize to both ORF probe sets, causing signal from one ORF to be reflected in both, whereas this would not arise in the cRNA case. This could be sufficient for data gathered from the same sample RNAs using the two different protocols to segregate into different clusters.
Establish Standards for Reporting Data that Cover All RNA Expression Assays
On the basis of issues that arose in generating the ERA file, we propose that researchers publish versions of data files with the following characteristics:
- 1.
- Data are reported at the FDR level. Here this proved non-trivial for SAGE. Our ExpressDB representation of SAGE data in terms of minimum and maximum SAGE tag counts provides one example of how FDR level reporting may be accomplished for methodologies where the entities for which experimental data is collected may not be uniquely assignable to the functional RNA units chosen for data reporting (see Methods). Corresponding uncertainties mentioned above about cross-hybridization of paralogs for Affymetrix and microarray experiments affect the accuracy of FDR-assigned values and may ultimately be addressed by calibrating hybridization with known family members.
- 2.
- Expression data is reported by publicly recognized stable identifiers for FDRs rather than names (in this case SGDIDs vs. ORF names). This would avoid the need for name resolution when combining data from different sources.
- 3.
- Data values considered to be in error (frequently reported in microarray data) are excluded, and multiple nonerroneous values are consolidated into a best estimate. This makes it easier to compare data sets and also easier on the database itself. Maintenance of multiple data values for an FDR in a condition incurs substantial database overhead and can lead to unexpected results in processing queries (see the hotlink for Multiple ORF Rows in the help document available from the EXD web query application). Ideally, experiments should be replicated and both central tendencies and variances of best estimates should be reported. This could have been done here for the Hol group of experiments.
- 4.
- Provide clear documentation on all reported measures describing their data sources, formulas used to compute them, and their proper usage.
- 5.
- Provide clear documentation on strains and environmental conditions for all data.
Some of these suggestions reaffirm the straw man standards of (Bassett et al. 1999) and, we hope, provide some concrete directions for following them. We emphasize that we do not suggest that this should be the only version of data that is published, but that such a version be prepared for to support easy comparison with other data sets. Availability of less processed forms will allow other researchers to explore error thresholds, characteristics of different probes for an FDR, etc.
As we noted previously, improvements in data comparability through establishment of standards for expression data collection, preparation, and reporting will make databases more useful. We emphasize that this pertains not just to ExpressDB but to any RNA expression database as the fundamental issue concerns limitations on the ability to compare data meaningfully, not the computer structures by which it may be stored and managed. Such improvements will also help streamline and focus databases. Taking ExpressDB as a case in point, in the absence of such standards, ExpressDB has both too much and too little data. On the too much side, a large number of the 2503 Measure records defined in ExpressDB and several million associated Expression Data Point (see Fig. 1) records are of little general scientific interest. They cannot be ignored because they are sometimes found to be essential to interpreting the data (e.g., microarray spot quality indicators); from there, general unclarity about data fields and their potential use, plus often sketchy documentation that makes it hard to distinguish potentially important from likely unimportant fields, offers no practical alternative to loading all reported data. On the too little side, information that is critical to interpreting expression profiles, especially strain and condition descriptions, is maintained in ExpressDB only in unformatted text. As functional genomics develops, a database will be required that maintains strain and condition information in a structured form that can be queried precisely for such characteristics as the presence of particular alleles in the strain, certain compounds in the medium, or treatments of the cell culture (e.g., heat shock). Moreover, ExpressDB's indexing of data at the RNA level, currently being generalized from ORFs to FDRs, will require further generalization. Not only are protein levels now being gathered on a high throughput basis (Link et al. 1997; Futcher et al. 1999; Gygi et al. 1999a,b; Page et al. 1999), but functional data on genomic features that do not generate RNA may need to be gathered, such binding affinities of proteins to regulatory DNA (Wang and Church 1992;Tavazoie and Church 1998). Phenotype information such as growth rates of mutant strains will also be of interest. To record such data will require indexing by proteins (including modified proteins), regulatory DNA, and strains in addition to FDRs. We have developed a logical model and some prototype components of a Biomolecule Interaction Growth and Expression Database (BIGED) that will enable many of these data to be integrated (J. Aach and W. Rindone, unpubl.). The logical model may be downloaded from our web site. ExpressDB and BIGED represent different tradeoffs between flexibility and biological meaning. Whereas ExpressDB flexibly allows any Measure cited in a load file to be recorded on the database regardless of its relation to and comparability with any other Measure, at the cost of their unstructured proliferation, BIGED defines structures for particular measures and biological features and relationships between them that correspond to their biological meanings, but requires more standardization of data contents and formats. We believe that both kinds of databases will be required as functional genomics advances.
METHODS
Database Model and Definitions
The ExpressDB database model and definitions were generated using PowerDesigner 6.1 (PowerSoft, Concord, MA). The full model is available at our web site http://arep.med.harvard.edu/ExpressDB/.
Database Loading
EDBUpdate was written in Perl and accesses the database using the sybperl interface. (See http://www.mbay.net/∼mpeppler/ for information on sybperl.) We edited load files where necessary using text editors and Perl scripts to put them into the required tab-delimited format with ORF names in a dedicated column. ORF names were converted to upper case. In some cases, we eliminated records from load files that could not be identified as representing either ORFs or controls. To enhance queriability of the data, we converted empty column positions in ORF rows (null values) to non-null default values, where meaningful defaults could be clearly identified; otherwise, null values were loaded as null database fields. To load SAGE data from Vel, we located SAGE tag sequences in yeast genome sequence downloaded from SGD and assigned them to ORFs on the basis of SGD ORF tables (both sequence and tables downloaded February 15, 1999) following rules for ORF and strand matching from (Velculescu et al. 1997). Because SAGE tags could not always be assigned uniquely to a single ORF, we computed and loaded minimum and maximum counts for each ORF as described in the text.
EXD Query System
EXD is a collection of Common Gateway Interface (CGI) (Gundavaram 1996) modules written in Perl with the sybperl interface. Some EXD CGI programs generate Javascript 1.2 code.
Generation of ERA File
Here we report methods used for generation of Affymetrix and SAGE ERAs; we discuss generation of microarray ERAs on our web site. We extracted average PM–MM values (Affymetrix) and ORF tag counts (SAGE) from ExpressDB for all Affymetrix and SAGE data sets listed in Table 1, along with any relevant qualifiers (e.g., Affymetrix feature set identifiers). We used a specially written batch extract routine for all Affymetrix database extracts; for SAGE data, we used EXD to extract only those ORFs for which counts were entirely unambiguous (minimum count = maximum count) for all conditions and only considered counts of 1 or more. We applied a standard sequence of processing steps to each individual load file, with variations as appropriate, to handle name and SGDID resolution, multiple ORF value consolidation, and Affymetrix threshold processing.
We standardized ORF names and assigned SGDIDs with a program that matched load file ORF names against an extract of the Name table of a prototype version of BIGED (J. Aach and W. Rindone, unpubl.; see web site), which had been loaded with all primary and alternate ORF names and all associations between ORF names and SGDIDs published on the SGD database since August, 1998. Name resolution also detected and matched hyphenation variants for some ORF names. The name resolution program added additional columns to the output name-resolved files that preserve an audit trail of the different names consolidated to their target standardized names.
For Affymetrix-derived files, the aggregate measure of the ratio of exon 1-based probe set values to exon 2-based probe set values mentioned in the text was the outlier-excluded average, over all ORFs with both exon 1 and exon 2 probe sets, of the ratio, for each such ORF, of the average over all conditions (n = 42 for Hol,n = 17 for Cho), of the average PM–MM values from an ORF's exon 1 probe set, to the corresponding average for the ORF's exon 2 probe set. We consolidated Affymetrix-based multiple probe set values for an ORF by examining Affymetrix probe set names and averaging the values of whichever group of an ORF's probe sets came first in the following sequence: (1) the probe set name is simply a gene name unqualified by exon or special feature set indicator (_i, _r, _f), (2) the probe set name is a gene name with an exon 2 designation with no special indicators, (3) the probe set name is a gene name with an exon 1 designation and no special indicators, (4) any other probe sets. Affymetrix probe set indicators such as _i, _r, and _f indicate that a probe set departs from desirable target rules for oligonucleotide probe sequence or probe set selection (Affymetrix Technical Help Desk, pers. comm.). The Rot Affymetrix-derived load file had already consolidated multiple ORF values and was exempted from this consolidation step.
Following multiple ORF value consolidation, Affymetrix-derived files with the exception of Rot were threshold adjusted to remove negative average PM–MM values. By and large, these correspond to ORFs considered absent by Affymetrix software (Lockhart et al. 1996); however, absence indicators were not available for all load files and we ignored them generally in favor of threshold adjustment. We performed threshold processing by computing, for each condition, the fifth percentile (P 5) of all consolidated ORF average PM–MM values, and replacing any consolidated ORF average PM–MM values for the condition <P 5 with theP 5 value; however, except for Cho many of theP 5 values were still negative and we used the fifth percentile of all positive values for them instead.
We collated all individual normalized load files into a single file using the standardized ORF names, combining all name audit trail information from each individual file. This file contained cleansed intensity and SAGE count values for each ORF for each experiment, but intensities are still on different scales for each condition. Finally, we produced a consolidated ERA file by dividing each non-SAGE-derived ORF intensity value by the total intensity value computed for its column; for SAGE-derived values, we computed ERAs by dividing each ORF count by the total number of non-rejected SAGE tags counted for the condition (Velculescu et al. 1997).
Condition Cluster Analysis
We transformed ERA data in preparation for clustering using a variant of procedures in (Tavazoie and Church 1998). For the entire collection of 217 conditions for which ERAs were computed (including microarray ERAs), we converted non-null ERAs for each ORF in each condition to log10 values, first adding the small value 0.000005 (equal to half the minimum precision non-zero value in the file) to all non-null values to eliminate zero values. We used logarithms to assure that induction and repression were on the same scale in so far as these are assessable through fold changes across conditions. If an ORF with expression level l in condition 1 is induced at fold change f>1 in condition 2 and repressed at the same fold change level in condition 3, then the difference between levels in 2 and 1 versus 1 and 3 isl(f − 1) versusl(1 − 1/f), and whereas the former term grows linearly with f and is unbounded, the latter term is hyperbolic and bounded within the interval (0,l). Because both correlation coefficients and direct clustering are based on difference terms, direct use of estimated relative abundance values risks underrepresenting the effects of repression (see also Eisen et al. 1998).
We then converted non-null log10 relative abundances for each ORF to standard units across all conditions to generate standard unit log10 relative abundances (SULRA). We performed the clustering of Figure 3 on Pearson correlation coefficients over ORF SULRAs between all pairs of our 217 conditions. However, many ORFs have relative abundances at or below the level of measurement noise and variation of their SULRAs over conditions can be expected to reflect noise as much as change in expression level. Also, some ORFs with higher relative abundance varied so little across conditions that difference terms between condition levels could also reflect noise. We therefore considered subsets of ORFs exhibiting high relative abundance levels and evidence of significant induction or repression as defined by two criteria: (1) the median ERA of the ORF over all conditions >= a percentile threshold p of the median ERAs of all ORFs, and (2) at least 10% of ratios of ERAs for the ORF over all pairs of conditions >= a threshold r, in which this latter was evaluated by ensuring that the ratio of thekth largest and kth smallest relative abundance level for the ORF ≥ r, wherek = ceiling[sqrt(n(n−1)/20)], wheren = number of conditions with non-null relative abundance values. We looked for subsets with ∼1000 ORFs. We used a subset of 1078 ORFs selected with P = 60 and r = 3 for Figure 3. Correlation coefficients were clustered using trace clustering (Ward's algorithm) in SPLUS 4.5 (MathSoft, Seattle, Wa.) We color coded the diagram using in Excel 97 (Microsoft, Redmond, Wa.)
Acknowledgments
We thank Pat Brown, Paul Spellman, Vishwanath Iyer, Joseph DeRisi, Michael Eisen, and Rick Young for their assistance in helping us understand data and procedures from experiments included in this analysis. Within the Church Laboratory, we thank Barak Cohen for use of unpublished data, and Barak Cohen, Rob Mitra, Saeed Tavazoie, Martin Steffen, and others, for helpful discussions on data cleansing, analysis, clustering, and for critical comments on this manuscript. We also thank four anonymous reviewers for very helpful critical comments. Finally, we thank the Lipper Foundation, Hoechst Marion Roussel, DOE grant DE-FG02-87ER60565, and Howard Hughes Medical Institute for their funding of this work.
NOTE ADDED IN PROOF
The update of the design of ExpressDB to be organism-independent, mentioned above, is now complete. Details on the new design are on our web site.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL church{at}salt2.med.harvard.edu; FAX (617) 432-7663.
-
- Received July 21, 1999.
- Accepted February 16, 2000.
- Cold Spring Harbor Laboratory Press