Holding Your Own: Establishing Sister Chromatid Cohesion
Chromosome Segregation and Genome Integrity
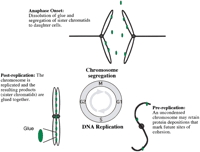
At cell division, both daughter cells receive only one copy of the parental genome. Not surprisingly, chromatin plays a role at numerous stages to ensure that genome segregation occurs with high fidelity. First, each chromosome is duplicated to produce two identical chromosomes called sister chromatids. Second, sister chromatids become paired, or glued together, along the entire chromosome length (Sumner 1991; Selig et al. 1992; Guacci et al. 1994). This pairing, or cohesion, provides a mechanism by which the identity of sister chromatids can be maintained over time. Third, sister pairs associate with the mitotic spindle apparatus via kinetochores, specialized protein complexes that assemble on centromeres. This association produces a highly defined geometry: The kinetochore of one chromatid associates with microtubules from the spindle pole opposite that of her sister. Cohesion between sister chromatids facilitates this orientation, most probably by physically constraining sister kinetochores to face in opposite directions (Rieder 1982; Waters et al. 1996; Michaelis et al. 1997; Tanaka et al. 2000a). Fourth, kinetochores associated with spindle microtubules generate poleward-pulling forces that act to separate sister chromatids (McNeill and Berns 1981; Rieder et al. 1986; Skibbens et al. 1995). Cohesion opposes these kinetochore pulling forces, stretching the intervening chromatin and generating tension across the chromatid pair. This tension is critical for coordinating sister chromatid movement during mitosis (Skibbens et al. 1995), and for cell cycle progression. In support of the latter, kinetochores capture microtubules by chance, such that initial attachments are not always correct (Nicklas 1997). Fortunately, cells have evolved a mitotic checkpoint mechanism that halts cell cycle progression in response to inappropriate kinetochore–microtubule interactions or in the absence of tension (Hoyt et al. 1991; Li and Murray 1991; Nicklas et al. 1995; Li and Nicklas 1997; Waters et al. 1998, 1999; Skibbens et al. 1999). Only after all of the chromatid pairs are properly oriented to the mitotic spindle apparatus does dissolution of cohesion occur, allowing sister chromatids to segregate away from each other and move into the newly forming daughter cells (Fig. 1).
Numerous advances have facilitated the molecular dissection ofcis and trans factors required for establishing sister chromatid cohesion. A seminal finding that propelled forward our understanding of cohesion and cell cycle regulation was that anaphase onset (sister separation) and exit from mitosis (M-phase to interphase transition) are separable events: The dissolution of sister chromatid cohesion could now be studied in an isolated context (Holloway et al. 1993; Surana et al. 1993; Irniger et al. 1995). Other advancements, including the adaptations of fluorescent in situ hybridization (FISH) and GFP-tagged repressor/operator methodologies, allowed for the visualization of individual sister chromatid loci in yeast cells, an organism in which mutants are readily generated and the genome sequence is known (Guacci et al. 1994; Straight et al. 1996; Michaelis et al. 1997; Megee and Koshland 1999). Finally, analyses of DNA from coimmunoprecipitates, in combination with functional assays of cohesion in minichromosomes, have provided a vehicle to identify DNA sequences that participate in cohesion (Dedon et al. 1991; Meluh and Koshland 1997; Blat and Kleckner 1999; Megee and Koshland 1999; Megee et al. 1999; Tanaka et al. 1999).
In this review, we take a cerevisiae-centric view of the recent advances that have brought insights, as well as intrigue, to the mechanism by which cis and trans factors participate in cohesion establishment.
Structural Cohesins
In budding yeast, several structural cohesion proteins (or cohesins) have been identified: Smc1p, Smc3p, Mcd1p/Scc1p, and Irr1p/Scc3p (Strunnikov et al. 1993; Kurlandzka et al. 1995; Guacci et al. 1997;Michaelis et al. 1997; Toth et al. 1999). These cohesins are conserved across evolution and their characterizations have been summarized in numerous reviews (Hirano 1999; Nasmyth 1999; Strunnikov and Jessberger 1999). Briefly, cohesin proteins are thought to function as structural components in sister chromatid cohesion based on three criteria. First, each protein is required for cohesion such that loss of function in any one results in sister chromatid separation before anaphase onset (Guacci et al. 1997; Michaelis et al. 1997; Toth et al. 1999). All loci separate in cohesin mutant cells, indicating that the mechanism of cohesion is conserved along the chromosome length. Note that factors unrelated to cohesins have been identified from other organisms that localize to, or function in, cohesion only at centromeres (Saitoh et al. 1997; Moore and Orr-Weaver 1998; Tang et al. 1998). Second, cohesins are nuclear proteins that associate with chromatin from S-phase through the bulk of mitosis: That portion of the cell cycle when sister chromatid cohesion must be maintained (Guacci et al. 1997;Michaelis et al. 1997; Toth et al. 1999; Ciosk et al. 2000). Data from yeast and Xenopus support a model in which all four cohesins function as a complex (Guacci et al. 1997; Michaelis et al. 1997;Losada et al. 1998; Toth et al. 1999). Third, cohesins dissociate from chromatin at anaphase onset when sister chromatids separate (Guacci et al. 1997; Michaelis et al. 1997; Toth et al. 1999; Uhlmann et al. 1999), although the kinetics of Smc1p and Smc3p dissociation are delayed relative to those of Mcd1p/Scc1p and Irr1p/Scc3p (Tanaka et al. 1999; Ciosk et al. 2000). In summary, these results are consistent with a model in which a complex required for cohesion associates with chromatin during the period of cohesion maintenance.
Cohesin dissociation from chromatin has been interpreted as a biochemical marker for anaphase onset and sister separation. For Mcd1p/Scc1p, dissociation is ultimately regulated by the anaphase-promoting complex (APC). APC, in association with the APC cofactor Cdc20p, targets Pds1p (an inhibitor of anaphase that exists in a complex with Esp1p) for degradation (Cohen-Fix et al. 1996; Schwab et al. 1997; Visintin et al. 1997; Ciosk et al. 1998). Pds1p degradation frees Esp1p to function as a positive regulator of anaphase onset. In yeast, Esp1p facilitates the two-site cleavage and dissociation of the cohesin Mcd1p/Scc1p from chromatids, possibly via an endopeptidase activity (Ciosk et al. 1998; Uhlmann et al. 1999; Nasmyth et al. 2000). In contrast, little is known concerning the mechanism and extent of Smc1p and Smc3p dissociation. Several reviews provide various details concerning anaphase onset and the dissolution of cohesion (Farr and Cohen-Fix 1999; Nasmyth 1999; Peters 1999).
Sites of Cohesion
With cohesion proteins in hand, researchers focused on identifying the DNA sequences that participate in cohesion. Such an analysis is daunting, not only because of the genome size of most organisms, but because it was unknown whether cohesins associated with (1) all DNA sequences, (2) specific sequences repeated throughout the genome, or (3) specific sites but in a sequence-independent manner. Numerous approaches were used to address this issue. Chromatin immunoprecipitation (CHIP) tests for cohesin deposition at both endogenous chromosomes and at ectopic DNA sequences inserted within yeast chromosomes or minichromosomes (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999). While CHIP is a powerful approach and has been used to identify cohesion sequences at relatively high resolution (200–300 bp), this method relies on signals obtained from a population of cells. In addition, cohesin subunits participate in different complexes to perform a variety of tasks (e.g., DNA recombination/repair, chromosome condensation, and gene expression), requiring the performance of other assays showing that cohesin deposition participates functionally in cohesion (Jessberger et al. 1996; Guacci et al. 1997; Donze et al. 1999). To produce an overview of cohesion deposition along an entire chromosome, the CHIP method was modified such that radio-labeled DNA obtained from chromatin coimmunoprecipitations was used to probe membrane filters that contained DNA fragments spanning chromosome III (Blat and Kleckner 1999). Thus, the sites and spacing of cohesion deposition were obtained for a whole chromosome, albeit with reduced resolution. In addition to CHIP-based methodologies, Megee and Koshland (1999) exploited the small size of yeast plasmids to minimize cohesion binding-site redundancy and induce recombination events to regulate in vivo excision of highly defined DNA sequences from plasmids. Sister chromatid dissociation of the resulting recombination products could then be directly monitored using FISH and GFP-tagged minichromosomes, thus providing a functional assay for cohesion on a per cell basis (Megee and Koshland 1999; Megee et al. 1999). In close agreement, the centromere was identified as a predominant cohesin-binding site (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999), consistent with cytological studies from higher eukaryotes in which sister chromatid centromeres are tightly paired and often not individually resolvable (Sumner 1991).
What portion of the centromere is required for cohesion? The yeast centromere is composed of three centromere DNA elements (CDE I, CDE II, and CDE III) that together comprise approximately 125 bp (Hyman and Sorger 1995). While the full-length centromere functions in cohesion and is a substrate for Mcd1p/Scc1p deposition, a single base pair substitution in CDE III significantly decreased Mcd1p/Scc1p deposition and cohesion between. CDE III allowed for only very limited cohesion binding and did not function as a cohesion site until a portion of CDE II was included. These results reveal that cohesion at the centromere is directed by as little as 100 bp that predominantly requires CDE III (a portion of CDE II followed by all of CDE III) (Megee and Koshland 1999; Megee et al. 1999; Tanaka et al. 1999). CDE III is also essential for chromosome segregation and centromere structures associated with kinetochore assembly (Hyman and Sorger 1995). Does cohesion rely on kinetochore function as opposed to centromere sequence? Tanaka and coworkers (1999) found that mutations in kinetochore proteins, including Ndc10p, Cse4p, and Mif2p, abolish Mcd1p/Scc1p deposition at ectopic centromeres (deposition at endogenous centromeres was only minimally affected). These results suggest that Mcd1p/Scc1p is recruited to, and participates in, cohesion at yeast centromeres but that this recruitment relies on kinetochore function, not centromere sequence.
In addition to the centromere, numerous chromosome arm loci have been identified as sites of cohesin deposition or cohesion (Blat and Kleckner 1999; Megee and Koshland 1999; Megee et al. 1999; Tanaka et al. 1999). Using a functional assay for cohesion, inclusion ofURA3 was found to partially compensate for reduced minichromosome pairing upon centromere excision (Megee and Koshland 1999). URA3 and LEU2 were also found to function as sites of Mcd1p/Scc1p deposition at their endogenous contexts (chromosomes V and III, respectively) (Megee and Koshland 1999; Megee et al. 1999). Other deposition sites for chromosome V include a region encompassing DMC1 and another just downstream fromYER184C (Tanaka et al. 1999). A site of cohesin deposition also has been identified proximal to the 5S gene within each rDNA repeat on chromosome XII (S. Laloraya and D. Koshland., pers. comm.). Although all of these sites have been resolved to within a few hundred base pairs, a motif that directs cohesin binding has not been identified, nor has a correlation of deposition to intergenic or intragenic loci been obtained. To date, the best correlation between cohesin deposition and DNA sequence resides in nucleotide bias: Chromatin regions that bind cohesins are high in A/T content, including the CDE II region of the centromere. Regions that are high in G/C content appear to demarcate cohesin association, possibly acting as a barrier to cohesin spreading (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999).
Given that cohesion between sister chromatids must be maintained through a significant portion of the cell cycle, it is surprising to find evidence that the cohesin-chromatin interaction is quite dynamic. First, ectopic insertion of a centromere into a site previously devoid of detectable Mcd1p/Scc1p levels results in deposition not only at the centromere, but also into flanking cold sequences (Megee et al. 1999;Tanaka et al. 1999). Cohesin spreading over time may help explain the robust and expansive signal observed for centromeres relative to arm sites (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999). Arm sites may also be competent to direct the spread of cohesin: When a region of high Mcd1p/Scc1p deposition was divided into eight short DNA sequences, only two of these sequences allowed for deposition when integrated to a new locus (Tanaka et al. 1999). Note that the inability of the remaining six segments to direct binding could instead relate to an inhibitory effect emanating from the flanking sequences (see below). Second, cohesin spreading occurs symmetric to flanking sequences but can also spread in an asymmetric fashion, indicating that flanking sequences can facilitate or inhibit cohesin spreading (Megee et al. 1999). Third, cohesins that spread to sequences adjacent to the centromere dissociate from those regions upon excision of the centromere, even when the centromere is excised as late as metaphase (Megee et al. 1999). Thus, cohesin's association with DNA must be actively maintained (at least at sites of cohesin spreading). Finally, cohesion must accommodate chromosome condensation. Cytological studies reveal that human chromosomes in late prophase appear as cylinders with unresolved sister chromatids, suggesting that a significant volume of sister chromatids has cohesion. As condensation proceeds, the individual sisters become more defined until, late in metaphase, each chromatid is highly resolved and paired along a narrowed axial region (Sumner 1991). This axis of cohesion does not simply appear to be the condensed sum of all the previous cohesion sites, because cells maintained in metaphase eventually lose cohesion along the arms. These results suggest that the majority of cohesion sites are lost before metaphase, consistent with studies performed in Xenopus andSordaria (Losada et al. 1998; van Heemst et al. 1999). Insight as to how the dynamic process of cohesion and condensation may coexist was obtained by the finding that cohesion factors Mcd1p/Scc1p, Ctf7p/Eco1p, and CHL12/CTF18 are required for aspects of chromosome condensation (Guacci et al. 1997; Skibbens et al. 1999) (J. Hanna et al., pers. comm.). Further study is required to determine the extent to which cohesion can be remodeled in response to DNA replication/repair, condensation, transcription, or other chromatin-altering processes.
Establishing Cohesion
The mechanism by which the products of DNA replication are identified as sisters remains elusive. Three factors, Ctf7p/Eco1p, Scc2p, and Scc4p, have been identified as acting early in the process of cohesion (Skibbens et al. 1999; Toth et al. 1999; Ciosk et al. 2000). Scc2p and Scc4p appear to function in cohesin deposition. First, Scc2p (Mis4p in S. pombe) (Furuya et al. 1998) and Scc4p combine to form a complex and function during S-phase when cohesion is established. Second, the structural cohesins, Mcd1p/Scc1p and Irr1p/Scc3p, fail to stably associate with chromatin in scc2or scc4 mutant cells (Toth et al. 1999; Ciosk et al. 2000), consistent with a deposition function for the Scc2p-Scc4p complex. The molecular mechanism by which this deposition occurs is unknown. For instance, while cohesin and deposition (Scc2p and Scc4p) complexes associate with chromatin, they infrequently colocalize and do not stably associate with one another, at least not in stoichiometric measures (Toth et al. 1999; Ciosk et al. 2000).
One model put forward in the literature posits that structural cohesins are preassembled into a soluble complex and then deposited onto the chromatin (Tanaka et al. 1999; Toth et al. 1999; Ciosk et al. 2000). For instance, all four cohesins physically associate to form a soluble complex and have been shown to coassociate with at least one discrete chromosomal locus (Tanaka et al. 1999; Toth et al. 1999). Further evidence for the requirement of preassembly before deposition is that Scc3p fails to associate with chromatin in cells harboring mutations in either MCD1/SCC1, SMC1, or SMC3 (or the deposition factor SCC2) while Mcd1p/Scc1p fails to associate with chromatin in cells harboring mutations inIRR1/SCC3 (or SCC2). A chromatin association for Mcd1p/Scc1 was not detected in SMC1 or SMC3mutant cells (Toth et al. 1999). These findings have been interpreted to show an interdependence of cohesin subunit association with chromatin: Deposition occurs in the form of an intact cohesin complex.
Formally, an alternate model is that cohesin deposition occurs by the stepwise addition of cohesins. It has been well established that Mcd1p/Scc1p is upregulated and begins to associate with chromatin in late G1/early S-phase (Guacci et al. 1997; Michaelis et al. 1997; Toth et al. 1999). However, Smc1p associates with chromatin before Mcd1p/Scc1p is upregulated and before Mcd1p/Scc1p or Irr1p/Scc3p associate with chromatin (Ciosk et al. 2000). That SMC protein association with chromatin may be the first step in cohesin deposition makes sense: Only SMC family members (shown for various constructs of cohesins Smc1p and Smc3p and condensin Smc2p) have been shown to directly bind DNA. Smc1p and Smc3p C-terminal fragments preferentially bind DNA substrates that are A/T rich or contain secondary structures. Of interest is that yeast centromere DNA, a prominent cohesion site that is A/T rich and contains inverted palindromic repeats, is one of many efficient competitors of Smc1p substrate binding (Akhmedov et al. 1998,1999). Eukaryotic SMCs most likely assemble into heterodimers (Hirano 1998; Jessberger et al. 1998; Nasmyth 1999). Note that bacterial SMC proteins are singly represented in the genome and thus assemble into homodimers (Melby et al. 1998). Thus, a likely scenario for eukaryotes is that an Smc1p-Smc3p complex associates with DNA, and that this deposition allows for the subsequent loading of Mcd1p/Scc1p and Irr1p/Scc3p. In Drosophila, the cohesion factor, MEI-S332, associates with chromatin after S-phase, providing evidence across evolutionary lines that cohesion between sister chromatids may occur in a multistep fashion (Kerrebrock et al. 1992; Tang et al. 1998).
Surprisingly, the combined activities of deposition (Scc2p and Scc4p) and cohesin (Mcd1p/Scc1p, Irr1p/Scc3p, Smc1p, and Smc3p) factors are still insufficient to establish cohesion. For instance, studies in yeast and Xenopus reveal that cohesins can bind chromatin (presumably via the Scc2p-Scc4p complex) before, during, after, or in the absence of DNA replication. However, when cohesin deposition is excluded from S-phase, sister chromatid cohesion is abolished (Losada et al. 1998; Uhlmann and Nasmyth 1998). Characterization of Ctf7p/Eco1p unveiled a third class of cohesion factors: those that function in cohesion establishment. Like SCC2 and SCC4,CTF7/ECO1 is required only during S-phase. However,CTF7/ECO1 mutant cells contain separated sister chromatids even though cohesin proteins associate with chromatin and appear properly targeted to cohesion sites (Skibbens et al. 1999; Toth et al. 1999; R. Skibbens and D. Koshland, unpubl.). Fundamentally, cohesion establishment, as mediated by Ctf7p/Eco1p, must include a mechanism in addition to cohesin assembly and deposition.
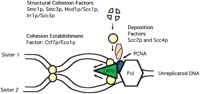
The finding that CTF7/ECO1 interacts genetically withPOL30 and CHL12/CTF18 provided one of the first molecular clues that the processes of DNA replication and cohesion establishment are intimately coupled (Skibbens et al. 1999).POL30 encodes the DNA polymerase cofactor proliferating cell nuclear antigen (PCNA) (Bauer and Burgers 1990). PCNA trimers form a tauroidal sliding clamp that associates with and promotes DNA polymerase processivity at the replication fork (Kelman 1997). PCNA is loaded onto dsDNA by a replication factor complex (RFC) (Cullmann et al. 1995; Jonsson and Hubscher 1997). Attesting to the specificity of the above interactions is that CHL12/CTF18 encodes an RFC-like protein (Spencer et al. 1990; Kouprina et al. 1993,1994). Because cohesion is established during S-phase (Sumner 1991; Selig et al. 1992; Guacci et al. 1994; Uhlmann and Nasmyth 1998; Skibbens et al. 1999; Toth et al. 1999), the sum of these results suggests a model in which cohesion establishment is directly coupled to DNA replication and takes place in association with PCNA at the replication fork where sister chromatids are closely apposed (Fig.2). This model is attractive because it helps explain how cohesion establishment ensures the specificity of chromatid pairing while at the same time precludes the catastrophic pairing of nonsisters or repetitive sequences within a chromatid. Mis4p, the fission yeast homolog of budding yeast Scc2p, also interacts with the DNA replication machinery (Furuya et al. 1998), providing support across evolutionary lines that cohesion establishment and DNA replication are coupled.
One possible mode of action for cohesion establishment. Structural cohesion factors Mcd1p/Scc1p, Irr1p/Scc3p, Smc1p, and Smc3p (yellow balls) are loaded as a complex or by stepwise addition onto chromatin via the Scc2p-Scc4p deposition complex (salmon rectangle). Deposition is shown to occur near the site of action of Ctf7p, but may occur anywhere before or after the replication fork. Establishment of sister chromatid cohesion requires Ctf7p/Eco1p (green triangle), which may move in association with PCNA at the DNA replication fork. Ctf8p, Ctf4p/Chl15p/Pob1p, Chl12p/Ctf18p, and the DNA polymerase Trf4p (Pol κ) are other cohesion factors that may associate with the DNA replication fork (not shown).
Cementing the link between DNA replication/repair and cohesion are two recent findings, the first that TRF4 encodes a DNA polymerase (Pol κ), which functions in sister chromatid cohesion (Wang et al. 2000). Pharmacological studies reveal that Pol κ (TRF4) shows sensitivities shared by B-polymerases, a class of DNA replication/repair enzymes (Pelletier et al. 1994; Wang et al. 2000). Whereas TRF4-TRF5 double mutant cells fail to complete DNA synthesis, TRF4 mutant cells complete the bulk of S-phase but contain a high frequency of separated sisters. It is not known how the Pol κ polymerase function of TRF4 facilitates cohesion, although TRF4 interactions with SMC1 have been reported (Castano et al. 1996). The second finding comes from fission yeast. ESO1, the fission yeast homolog ofCTF7/ECO1, has been isolated and is required for sister chromatid cohesion, showing that CTF7/ECO1 is conserved in function across evolution (Tanaka et al. 2000b). Intriguingly, ESO1 is much larger than its budding yeast counterpart and appears to be the fusion of budding yeastRAD30 as its N-terminus and CTF7/ECO1 as its C-terminus (Skibbens et al. 1999; Toth et al. 1999; Tanaka et al. 2000b). RAD30 encodes yet another DNA polymerase (Pol η) that functions in DNA repair (Johnson et al. 1999). RAD30function in cohesion has yet to be tested. Interestingly, mutations in one of the human homologs of RAD30 (Pol η) have been identified in patients afflicted with xeroderma pigmentosum, a disease associated with UV-induced oncogenesis (Woodgate 1999).
Good things can come in threes, and characterizations of the three newest cohesion factors (CTF8, CHL12/CTF18, and CTF4/CHL15/POB1) provide exciting insights into the mechanism of cohesion. CTF8, a chromosome transmission fidelity mutant, is synthetically lethal with known structural (MCD1/SCC1 and SMC3) deposition (SCC2) and establishment (CTF7/ECO1) cohesion factors. Given these interactions, it is not surprising thatCTF8 mutant cells are defective in sister chromatid cohesion (M. Mayer, S. Gygi, and P. Hieter, pers. comm.).CHL12/CTF18, implicated in cohesion by genetic interactions with CTF7/ECO1 (Skibbens et al. 1999), also functions in cohesion (J. Hanna, E. Kroll, V. Lundblad, and F. Spencer, pers. comm.). In a remarkable convergence of independent endeavors, numerous methodologies show thatCHL12/CTF18 and CTF8 interact with each other and with bona fide RFC components (RFC4 and/orRFC3) (J. Hanna, pers. comm.; M. Mayer, S. Gygi, and P. Hieter, pers. comm.). Note that while CHL12/CTF18contains a region conserved among RFC factors (and most notably,RFC1), CHL12/CTF18 does not appear to associate with the RFC complex (RFC1-RFC5) that functions in processive DNA replication (J. Hanna, pers. comm.). Previous studies showed that RFC components (RFC2-RFC5) combined with the DNA damage checkpoint protein RAD24p, which contains regions of homology with RFC1 (Ellison and Stillman 1998; Green et al. 2000). The interactions described above extend this finding and suggest that there may be several such RFC-containing complexes, one of which may scan along DNA in association with the DNA replication fork to function in cohesion.
Whereas CTF4/CHL15/POB1 has not yet been implicated in interactions with bona fide RFC components, this third-newest cohesion factor is required for cohesion, and genetically interacts with CHL12/CTF18 and several genes implicated in DNA synthesis, including DNA polymerase α (Miles and Formosa 1992; Wittmeyer and Formosa 1995; Formosa and Nittis 1999; J. Hanna, E. Kroll, V. Lundblad, and F. Spencer, pers. comm.). Of interest is that bothCTF8 and CHL12/CTF18interact with telomere metabolism genes and function in telomere length regulation (M. Mayer, S. Gygi, and P. Hieter, pers. comm.; J. Hanna, E. Kroll, V. Lundblad, and F. Spencer, pers. comm.). Can the mechanisms of cohesion be central to so many different processes (telomere metabolism, chromosome condensation, DNA replication), or are the various phenotypes shown by cohesion mutant cells a secondary consequence of cohesion loss?
Whereas the challenges that face researchers interested in sister chromatid cohesion still loom large, the summit is much closer and the trails are clear. Specific milestones that we will likely find along the way include discerning that portion of the cohesion machinery that resides at the DNA replication fork as opposed to that which resides and functions on discrete loci after fork passage. In addition, it will be interesting to determine how mediators of DNA topology (DNA replication, transcription, silencing, etc.) influence cohesion and site selection. Coming full circle, cohesion factors may themselves remodel chromatin to facilitate the site-selection process (Hirano and Mitchison 1994; Saka et al. 1994; Hirano et al. 1997; Sutani and Yanagida 1997; Losada et al. 1998; Kimura et al. 1999). For instance, mutations in numerous cohesion factors have been shown to functionally alter or abolish centromere structures or higher-order assemblies on chromosome arms (Spencer et al. 1990; Castano et al. 1996; Guacci et al. 1997; Saitoh et al. 1997; Goshima et al. 1999; Skibbens et al. 1999; van Heemst et al. 1999; J. Hanna, E. Kroll, V. Lundblad, and F. Spencer, pers. comm.). Finally, if the cohesion machinery scans the DNA to identify sites for cohesin deposition, how are these complexes regulated and for what does it scan?
Acknowledgments
The author thanks Drs. Meg Kenna, Vincent Guacci, Paul Megee, F. Spencer, M. Mayer, and laboratory members for their critical reading of the manuscript, their time, and helpful discussions. The author also thanks those researchers who kindly shared information before publication: S. Laloraya, V. Guacci, D. Koshland, J. Hanna, E. Kroll, V. Lundblad, F. Spencer, M. Mayer, S. Gygi, and P. Hieter.
Footnotes
-
E-MAIL rvs3{at}lehigh.edu; FAX (610) 758-4004.
-
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.153600.
- Cold Spring Harbor Laboratory Press