ACAPELLA-1K, A Capillary-Based Submicroliter Automated Fluid Handling System for Genome Analysis
Abstract
The Genomation Laboratory in the Electrical Engineering Department at the University of Washington has been developing an automated, high-throughput, submicroliter-scale fluid-handling system for use in molecular biology, especially as part of the Human Genome Project and other high-throughput DNA sequencing endeavors. Small glass capillaries enable the preparation, handling, and monitoring of 1-μl reaction volumes. The Genomation Laboratory, with corporate partners Orca Photonic Systems, Inc. and Engineering Arts, has developed modules for aspiration, dispensing, mixing, transport, and rapid thermal processing of biological samples contained in glass capillaries. The ACAPELLA-1K is the first integration of these modules, designed to process 1000 samples in an eight-hour day. It has served as a test bed for the technologies as well as for performing biological experiments in conjunction with the University of Washington Genome Center. This system and related results are presented in this paper. A video of the system in operation is provided at http://www.genome.org. The Genomation Laboratory is presently developing the next-stage ACAPELLA-5K system based on the results of the ACAPELLA-1K system.
Molecular cloning, enzymatic reactions, electrophoresis, fluorescence imaging, signal processing, and data handling are the basic methods of modern genomics research. Although many devices exist to automate these processes, sample handling within and between systems is not well integrated, has low throughput and high failure rates, and consumes larger volumes of expensive and precious materials than is required by current detection technologies. To reap the benefits of the information obtained from genome analyses such as large-scale DNA sequencing, diagnostics, and DNA fingerprinting more rapidly, increased throughput and decreased costs are essential.
Dramatically improved DNA sequencing technology can be developed through the application of miniaturization and automation to state-of-the-art genomic sequencing processes. Discrete instruments have been developed to automate nearly every step in the large-scale sequencing process but few address both miniaturization and full automation. At the Stanford DNA Sequencing and Technology Center (http://sequence-www.stanford.edu) a modular automated sequencing system is being developed with the goal of sequencing 1 Mb/day of assembled sequence at a cost of 1 cent per base. Highly automated systems are designed with a modular approach where samples are transferred manually from one device to another via a common interface, a cassette of fourteen 96-well microplates (Marziali et al. 1999). Other groups such as the University of Texas Southwestern Medical Center Genome Science and Technology Center (http://gestec.swmed.edu) and the Whitehead Institute/MIT Center for Genome Research (http://www-genome.wi.mit.edu) are working on automation of various aspects of the genome analysis process as described in D. Meldrum (in prep.). Most operations use a combination of manual methods, in-house automation, and commercial automation. Although significant advances have been made in automation in the past several years, few approaches utilize reduced fluid volumes or have modular processes that integrate well through a common interface.
A tremendous effort is being made by researchers to miniaturize genome analysis by developing technology for “lab-on-a-chip” systems (Harrison and van den Berg 1998). Adopting semiconductor manufacturing and plastic injection molding technologies to these systems promises to decrease costs significantly because of reduced manufacturing costs and miniature sample volumes. However, at present these lab-on-a-chip devices are not to the point that they are useful in most genome analyses settings, particularly large-scale DNA sequencing.
The development of ultrasensitive fluorescence-based electrophoresis systems for sequencing in both capillary [e.g., Bashkin et al. 1996;Kheterpal and Mathies 1999; Molecular Dynamics MegaBACE 1000, Sunnyvale, CA; Perkin Elmer (PE) ABI Prism 3700, Foster City, CA] and slab-gel formats (e.g., PE ABI 377 and Amersham Pharmacia Biotech ALF DNA sequencer, Piscataway, NJ; Smith et al. 1985; Hood et al. 1987) has already outstripped upstream sample-handling capabilities (Hunkapiller et al. 1991; Mathies et al. 1998). In addition, with the most sensitive detection systems, most of the sample is discarded because it is currently impossible to carry out the biochemical sample preparation steps in the small volumes that the electrophoretic and detection equipment are capable of sensing. By reducing the volumes of DNA samples and reagents, not only will there be a direct cost benefit for the sample preparation steps, but many of the upstream steps will benefit as well. For example, a change from 10 to 1 μl in the standard sample volume for the enzymatic reactions would change required culture volumes from milliliters to hundreds of microliters, thereby reducing the resources required for culture oxygenation, cell harvesting, and the fluid handling associated with automated DNA extraction protocols or procedures.
In the Genomation Laboratory (http://rcs.ee.washington.edu/GNL/genomation.html) at the University of Washington (UW) Department of Electrical Engineering, a fluid sample handling system called ACAPELLA-5K is being developed to process 5000 reactions of 1-μl each in 8 hr at a cost of <10 cents per finished sequence base pair. This is being accomplished through a 10-fold reduction in reagents and sample volumes and a 10-fold increase in throughput derived from automation and the handling of fluids in glass capillaries. The system is designed to interface in an automated fashion with 96- or 384-well microplates at the input end, and with electrophoretic capillaries, electrophoretic slab-gels, microplates, or capillary cassettes at the output. The system uses glass capillary tubes instead of high-capacity microplates to reduce sample size, automate the handling of small fluid samples, provide excellent volume control, scale from large to small sample volumes, access fluid from both ends, reduce thermal cycling times, minimize evaporation because of their low exposed surface-area-to-volume ratio, and minimize the amount of disposables used to perform DNA sequencing. Compared with microplates, capillaries have a higher heat exchange rate for faster thermal cycling and may be handled both sequentially and in parallel. Capillaries are also natural pipettes, ideally suited for removing fluids from microplates or other input sources and depositing samples onto gels or other output media. All processing steps are performed within these capillaries: preparation of the reaction cocktail (DNA and reagents are added), mixing of the fluids in the reaction, thermal cycling, and purification. In the ACAPELLA-1K system, all steps are automated except thermal cycling and purification. In the ACAPELLA-5K all steps will be automated. Although the primary target application for this system is large-scale DNA sequencing, it is a general-purpose chemical analysis system that may also be used for diagnostics, environmental testing, protein crystallography, mass spectrometry sample preparation, and so on.
Based on early proof-of-concept ideas (Meldrum 1997), the Genomation Laboratory, Orca Photonic Systems, Inc. (Redmond, WA) (http://www.orcaphoton.com), and Engineering Arts (Seattle, WA) (http://www.engineering-arts.com) have worked together to design, develop, and test the ACAPELLA-1K system capable of processing 1000 samples in eight hours. This first-generation instrument was developed to test technologies that will lead to development of ACAPELLA-5K, a system with higher throughput and additional funtionality. The ACAPELLA-1K instrument is described and the results presented in this paper. A video of the system in operation is provided at http://www.genome.org.
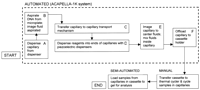
A detailed discussion of the ACAPELLA-1K subsystems is provided in Methods; here, a general overview of sample processing in the system (Figs. 1 and 2) is presented. Capillaries are loaded into a capillary dispenser (Figs. 1A and 2A) and DNA samples in a 96- or 384-well microplate are placed at the input station (Figs. 1B and 2B). With an aspirator-gripper, the system automatically takes a capillary from the capillary dispenser and aspirates an individual DNA sample into the glass capillary. The capillary is then placed onto a transport mechanism or comb (Figs. 1C and 2C) that moves the capillaries from one reagent dispenser to another. Piezoelectric reagent dispensers (Figs. 1D and 2D) dispense reagents into each capillary that moves along the transport comb. At the end of the transport comb, a mixer (Figs. 1E and 2E) mixes the sample and reagents. An imager at the input station (Figs. 1B and 2B) verifies the fluid volume aspirated and an imager at the mixer (Figs.1E and 2E) centers the fluid for mixing and verifies the total reaction volume. An off-loader takes the capillary from the mixer and loads it onto a capillary cassette at the off-loading station (Figs. 1F and 2F). The cassette is placed in a thermal cycler for performing the PCR, sequencing reactions, or restriction enzyme digests. The entire system is under computer control and has an intuitive, graphical user interface. All subsystems operate simultaneously as each capillary moves through the production line in a serial, pipeline fashion. For an analysis of the system throughput including serial versus parallel processing, see Meldrum (1997). The ACAPELLA-1K system aspirates DNA in volumes on the order of 100 nl ± 5%, dispenses reagents with a resolution of 300 pl ± 2%, and processes final reaction volumes of 1–2 μl. See Methods for a detailed description of ACAPELLA-1K and each one of its subsystems.
The ACAPELLA-1K automated fluid sample handling system. (A) Capillary dispenser; (B) input station (microplate with DNA samples, aspirator gripper, and imager); (C) transport comb; (D) piezoelectric reagent dispensers; (E) mixer and imager; (F) offloading station.
RESULTS
The ACAPELLA-1K system has demonstrated successfully that the automated preparation of 1500 reaction volumes of 1–2 μl each in 8 hr is possible even though it was designed for a throughput of 1000 capillaries. Experiments run on the system demonstrate the compatibility of the system with a variety of biological protocols (restriction digests, PCRs, and sequencing reactions) and that no cross-contamination is present. Through repeated use of the system, we have modified and improved it so that it is highly reliable and provides reproducible results.
Throughput and Reliability
The ACAPELLA-1K system has been used for fully automated reaction preparation of restriction digests and PCRs as well as semiautomated preparation of sequencing reactions. At present, it operates at a sample-handling throughput rate of >1500 PCR reactions of 2 μl each per 8-hr day. This throughput was determined experimentally. From September 1998 to September 1999, there have been seven runs of ∼1000 capillaries each on ACAPELLA-1K. Approximately 17,000 capillaries were run in the preceding year. Of 44,000 capillaries processed, approximately half were run for purposes of machine testing and half for various forms of chemistry and biology testing.
PCR reactions were prepared and analyzed in about 9000 capillaries. Several hundred sequencing reactions have been prepared and analyzed (30% dye primer and 70% Big Dye terminator). The remaining runs were primarily for testing and optimizing the system hardware. After initial debugging, operations have become quite smooth and reliable. Table1 provides a number of statistical measures of throughput and reliability acquired over the last 12 months of operation. The mechanical error rate, defined as events that lead to the loss of a sample capillary, result in undetectable product yield, or require human intervention has fallen to <0.5%. DNA sample aspiration from microplates has a 0% failure rate. Although a DNA sample is always aspirated from a well in the microplate, the volume varies ±5% over volumes from 100 nl to 1 μl. The most important factors for successful operation of the piezoelectric reagent dispensers are cleaning and choice of operating parameters (pulse shape and frequency). Priming of the dispensers requires 10 μl per reservoir. For reservoirs with a capacity of 1 ml, this is only a 1% increase in the volume so priming volumes do not significantly increase the cost of the reagents used. The piezoelectric mixer (Evensen et al. 1998) has been working flawlessly for the past 6 months. Losses during thermal cycling caused by incomplete sealing have recently been reduced to near zero with an improved sealing mechanism. There has never been capillary breakage from the sealing process.
Summary of ACAPELLA-1K Processing Results
PCR Results
More than 9000 PCR reactions have been automatically prepared, one per capillary on ACAPELLA-1K, and analyzed with an overall average failure rate of 1.57%. Product sizes successfully amplified range from about 200 bp to 1.5 kbp of human genomic DNA. A subset of the experiments is described below.
As part of the integration of the system into the production pipeline at the University of Washington Genome Center (UWGC), ACAPELLA-1K was used for PCR-based STS screening of BAC superpools (Asakawa et al. 1997). In a modification of the procedure described by Asakawa, 147,000 BAC clones, which provide 8× coverage of the human genome, are pooled into 48 superpools, each containing 3072 clones. After PCR screening identifies the superpools that contain the STS of interest, a second PCR is applied to 28 unique BAC pools prepared from three-dimensionally assigned clone groups of the particular superpool. A final round of PCR screening on the final four BACs identifies the clone of interest. For 8× BAC coverage of the human genome, 160,000 clones would need to be identified and ordered. Clearly, high-throughput automation would benefit this effort. Because the protocol was both simple and necessary, it was selected as a means of field testing the system.
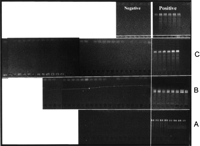
To evaluate the ACAPELLA-1K system and the use of 2-μl PCR reactions for this application, screens were performed on the 48 top-level superpools provided by the UWGC. Results from part of such a scan are shown in Figure 3. Four pools are identified as positives, matching the results of the UWGC. Significantly, no evidence of cross-contamination is seen, and the 2-μl reaction volumes are found to be adequate for these studies. Of course, some sites are difficult to amplify from the superpools and a brighter signal may be needed for the simple detection techniques employed here. For qualitative PCRs such as these, concentrating the sample into smaller wells can enhance the band intensity on the gel. A technique for doing this by using the glass capillaries for well formation and sample loading has been demonstrated (Evensen et al. 1999).
ACAPELLA-1K Superpool PCR screening with four positives shown. Reactions are 2 μl. Samples are human genomic DNA from the UWGC.
Reaction volumes <2 μl have been prepared. Figure4 shows the results of 1-μl PCRs performed on the plasmid vector pET21 (Novagen Inc., Madison WI), both with and without an insert. As shown, the small reaction volume produces an easily detected amount of product, despite the use of only 0.05 unit ofTaq DNA polymerase. As before, no evidence of cross-contamination between reactions with the two different templates is seen. The nonuniform results shown in Figure 4 are caused by loading variations.
ACAPELLA-1K 1-μl PCR of pET21 with and without insert (the break in the pattern is a gel loading error).
Reagent Stability Tests and Cross-Contamination Studies
An early concern in the development of the ACAPELLA-1K system was the viability of various biochemical reagents while being held in a dispenser reservoir for as long as eight hr at room temperature. As shown in Figure 5, we have failed to see any degradation in PCR product yield under these conditions.Figure 5A shows PCR product from the first 100 capillaries of a run of 1000 STS PCRs passed through the ACAPELLA-1K system, and Figure 5B shows product from the last 100 capillaries, which are processed over eight hr later. Both sets of samples used the same reservoir solutions of primers, enzymes, and proteins. This result is typical for day-long operation of the system. In the results presented, different gels were used to eliminate the chance of degrading the first set of samples while waiting 8 hr for the second set of samples.
(A) PCR results from the first 100 capillaries in one day's run of 1000 capillaries on ACAPELLA-1K; (B) PCR results from the last 100 capillaries in the day's run. Little if any degradation of the PCR components occurrs over the course of an 8-hr day.
Similar studies were performed to verify the stability of dye primer and dye terminator sequencing reaction reagents at room temperature. For PE Big Dye terminator reactions, the cocktails were prepared without DNA using the ACAPELLA-1K system. The cocktail was left at room temperature for 12 hr. After 12 hr, DNA was added to the mixture, the product was purified using Dynal magnetic beads and sequenced at the UWGC. The control experiment was exactly the same as above except that the reagents were not left at room temperature for 12 hr. Sequencing results from the experiments are nearly the same: A control sample had Phred quality scores of >20/646 and >40/340 with a read length of 1029; a sample with reagents left out for 12 hr had Phred quality scores of >20/648 and >40/461 with a read length of 1028. Similar experiments were run with Amersham Dye Primer reagents and successful results were obtained. Thus, the sequencing reagents used in the ACAPELLA-1K system are stable at room temperature for runs of 12 hr or less.
Another issue for the ACAPELLA-1K system and all automated devices used in molecular biology is the risk of cross-contamination. This concern drives the use of noncontact piezoelectric dispensers to distribute reagents to the individual reaction capillaries. Possible sources of contamination on the ACAPELLA-1K system could include DNA transfer via the capillary transport mechanism or aerosol contamination during mixing (Evensen et al. 1998). To date, no cross-contamination has been observed on the ACAPELLA-1K system in ∼15,000 PCRs. In an experiment to detect cross-contamination, amplifications of the polymorphic segments 5′ to the antithrombin III (ATIII) gene were performed on three DNA samples from the Centre du Etude Polymorphisme Humain (CEPH) library that are known to exhibit the three different allele combinations. As shown in Figure 6, no hint of cross-contamination is present. This was borne out over the course of the day and with a subsequent experiment described below.
Two-microliter reaction polymorphic PCRs on three different DNA samples prepared on ACAPELLA-1K.
To verify that there is no contamination in the ACAPELLA system, we performed a targeted cross-contamination study. Two runs of 600 capillaries and 1000 capillaries were run on different days. In each run, 96-well microplates were loaded with DNA samples of 6 ng/μl in the wells of columns one, four, and eight. The rest of the wells were loaded with water. Each well was aspirated once per cycle of 96 for a total of 6 times per well (run of 600 capillaries) or 10 times per well (run of 1000 capillaries). After preparing the reactions in the ACAPELLA system and thermal cycling the PCR products in the Idaho Technology air thermal cycler, the products were electrophoresed on a 2% agarose gel. A typical sampling of the results is presented in Figure 7. PCR products as expected resulted from the sample wells containing DNA and no PCR product resulted from the sample wells containing water. There were no errors (either false positive or false negative). Thus, there is no carryover to neighboring wells nor is there carryover of DNA from one DNA well to another DNA well. Overall, there is no detectable sample carryover in the ACAPELLA system.
Results of PCR cross-contamination study. There were 0 false-negative or false-positive results during 10 runs of an entire plate. The eight right-most lanes of A, B, and C were run with 1.5 ng/μl DNA, beginning from 6 ng/μl in the sample wells. The lanes to the left are samples processed with pure water from other wells of the same plate (16 lanes in A, 24 inB, 32 in C).
Restriction Enzyme Digest Results
Figure 8 shows the results of an automated, small-volume digestion of λ DNA with EcoRI (GIBCO BRL). The 2-μl reaction volume consists of 8.3 ng of DNA and 3.3 units of enzyme per capillary and 0.5 mg/ml BSA.
Sequencing Reaction Results
In addition to PCRs and restriction digests, we have also completed a number of semiautomated tests of full sequencing reactions, for both dye terminator and dye primer protocols. In a typical read from an ET-Dye Primer (Amersham) reaction of a UWGC human genomic DNA sample prepared on ACAPELLA-1K the results were quality > 20, 622; quality > 40, 115; and read length, 1158. In a typical read from a Big Dye Terminator (PE) reaction of a UWGC human genomic DNA sample prepared on ACAPELLA-1K the results were quality > 20, 569; quality > 40, 429; and read length, 1324. For both protocols we have obtained low failure rates, consistent read lengths of >1000, and Phred 20 scores of >500 bp. There were no failures for 200 samples analyzed. Purification of the dye terminator reactions before sequencing was performed manually with magnetic beads inside of 5-μl glass capillaries. See Methods for details. Development of a fully automated version of this purification procedure is in progress.
DISCUSSION
ACAPELLA-1K has successfully met the goals of preparing 1000 samples in 8 hr for restriction enzyme digests, PCRs, and sequencing reactions. The goals of the sequencing studies, to prepare capillary-based small-volume (1–2 μl) dye-terminator and dye-primer reactions that when sequenced yield read lengths and quality scores similar to those achieved by traditional methods, have been met. Automation of the purification stage is in progress. No evidence of cross-contamination exists in the system from the aspirating, dispensing, and mixing stations. Reagent stability tests have demonstrated that the reagents for PCRs and sequencing reactions are stable for 8 hr of operation at room temperature; thus, no refrigeration of reagents is required in the automated fluid handling system.
Although ACAPELLA-1K uses higher concentrations of polymerase and other reagents (see Methods) than are used in traditional 20 to 40-μl reaction volumes, it uses smaller amounts per reaction. These results are consistent with the findings of other researchers performing PCR reactions in small volumes (Cheng et al. 1996; Taylor et al. 1997). Higher reagent concentrations are most likely required because of limited diffusion in the elongated reaction chamber (Wittwer 1990;Kalinina et al. 1997) and the high surface-to-volume ratio of the capillary.
The ability to dispense small subnanoliter reagent volumes with the piezoelectric dispensers is a key to preparing microliter-to-submicroliter total reaction volumes. Although 10 μl of reagent is required to prime each piezoelectric dispenser, the reagent usage and cost is negligibly increased by 1% since reservoirs can hold 1 ml or more of reagents.
Based on the experience gained in developing the ACAPELLA-1K system, we have recently designed and built the ACAPELLA-5K system. This system reduces the number of capillary hand-offs, parallelizes the mixing stations, and combines the mixing and aspirating functions into one device. Initial experiments will begin in January 2000.
METHODS
The overall operation of ACAPELLA-1K is described in the introductory section and Figures 1 and 2. A detailed description of each one of the subsystems is included here.
ACAPELLA-1K Subsystems
Capillary Dispenser
A novel capillary dispenser (Fig. 9) holds 3800 capillaries and dispenses one capillary at a time (Sjoboen and Meldrum 1998). A newer modified version was recently built to hold 5000 capillaries and has been integrated with the ACAPELLA-1K system. It can be disassembled easily for cleaning if necessary. The dispenser is designed to store glass capillaries reliably in a known orientation, grab one capillary at a time in a groove on a slider, and positively insert one capillary on demand into a robotic gripper head. A unique feature of the geometry for the capillary storage unit keeps the capillaries from jamming. Currently the system uses Drummond glass capillaries or “microcaps” (Drummond Scientific Company, Broomall, PA) with a 5 μl capacity, 55 mm length, 838 μm outside diameter, and 340 μm inside diameter. The system is also able to accommodate the Drummond 2 and 8-μl glass capillaries as they have the same outer dimensions as the 5-μl glass capillaries.
Input Station
The input station (Fig. 10) consists of a microplate holder, a novel aspirator-gripper, and a CCD imager. Once a capillary has been pushed through the exit slot on the back of the capillary dispenser, a robotic aspirator-gripper grabs the capillary from the dispenser in a horizontal orientation, rotates the capillary down 90°, and moves it to the microplate to aspirate a DNA sample from a well specified by the computer. Currently, an operator must place the 96- or 384-well microplate on the input station by hand but future systems will include an automatic plate stacker. The aspirator-gripper can aspirate fluid volumes ranging from 100 nl to 1 μl ± 5% using the built-in piezo actuator. For U-bottom, 96-well microplates, each well must contain at least 2 μl for aspiration, whereas for conical-bottom microplates it is possible to empty the well with the aspirator. The CCD imager consists of a CCD monochrome camera (Edmund Scientific H53306) and software developed in-house to process the image. It is used to verify that the correct volume of fluid has been aspirated.
Input station for microplate of samples (A), aspirator (B) with capillary (C), and imager (above).
Transport Comb
Once a sample has been aspirated from a well of the input microplate and imaged, the robotic aspirator-gripper rotates the capillary up 90° in a horizontal orientation and places it onto the beginning of the transport mechanism or comb (Fig.11). This novel transport comb has three parallel sets of teeth and a grooved roller bar at the back. The inner pair of teeth rotate in a circular fashion to move the capillaries from one slot to the next on the outer set of teeth and the grooved roller bar. The outer pair of teeth do not provide a precision location for the capillary ends. An alignment notch, a component of the piezoelectric dispenser mount, is slightly higher than the tooth and locates the capillary at each one of the piezoelectric reagent dispensers. The grooved roller bar turns constantly, keeping the capillaries aligned against a back plate so that the opposite, wet end is held away from the reagent dispensers. Note that the end of the capillary that touches the plate does not touch any fluid so cross-contamination at the back plate is not an issue.
The capillary dispenser and aspirator-gripper continue to prepare capillaries with samples for the transport mechanism as other capillaries move down the transport comb for continued processing. This serial pipeline approach ensures that all subsystems are working simultaneously to process samples to meet the desired 1000 capillary per 8-hr day throughput. This serial approach also allows each sample to be processed with unique recipes if desired.
Piezoelectric Reagent Dispensers
Reagents are dispensed with piezoelectric ink-jet reagent dispensers (Fig. 12) designed and built by Engineering Arts (Seattle, WA) (http://www.engineering-arts.com). Droplets are dispensed in volumes of ∼300 pliters ± 2% from the reagent dispensers into the ends of the glass capillaries. The dispenser and the capillary do not touch, thus avoiding cross-contamination. Through the user interface, the user specifies the desired volume of each reagent to be dispensed. For example, if 0.3 μl is desired, then 1000 drops are dispensed from the dispenser.
Piezoelectric reagent dispenser (A) dispensing fluids into the end of a 340-μm inner diameter glass capillary (B) on transport comb (C) and aligned with the capillary indexer (D).
Mixer
A novel mixer actuator (Figs. 1E and 2E) and an optimized mixing protocol were designed to mix the fluids inside the capillary and to remove any bubbles contained in the fluids (Evensen et al. 1998). The fluid is moved back and forth by air volume displacement driven by a piezoelectric actuator. This actuator design is very similar to the aspirator-gripper. Rapid mixing of different fluids is achieved via diffusion between the main fluid volume in the capillary and the thin film it deposits on the capillary wall through its motion. Bubbles in the fluid are processed out of the capillary by use of an asymmetric velocity profile. In the experiments performed on the ACAPELLA-1K system, complete mixing takes anywhere from 3 to 10 sec in the 5-μl capillaries depending on the viscosity of the fluids and the volume of fluids being mixed. Experiments performed with λ DNA and BACs demonstrated no shearing of the molecules caused by aggressive mixing, and no evidence of aerosol contamination in PCRs has been found to date.
Before fluids are mixed with the mixer actuator, the fluids are viewed with a CCD imager and centered. Centering provides clearance between the fluid volume and the ends of the capillary for mixing. Imaging at this stage in the process is also used to monitor the total fluid volume in each capillary. The CCD camera and imaging software are the same as that used at the input station.
Off-loader Station
The off-loader station (Figs. 1F and 2F) consists of a robotic vacuum gripper and an output cassette. Once the sample and reagents have been mixed inside the glass capillary, the capillary is picked up by a vacuum gripper and placed onto a 32-capillary cassette. An operator picks up the loaded cassette and pushes the sides closed. The sides of the cassette are lined with translucent silicone rubber (McMaster Carr) that presses against the ends of the capillaries to seal them. At this point the loaded cassette is ready for thermal cycling, gel loading, or storage.
Thermal Cycling
Thermal cycling is performed with an Idaho Technology RapidCycler (Idaho Falls, ID). This cycler was designed for glass capillaries (Wittwer et al. 1989; Wittwer and Garling 1991) and can process 48 capillaries through 30 cycles in about 20–30 min. The ACAPELLA-1K 32-capillary cassettes fit into the three slots of the RapidCycler so 96 capillaries can be cycled at once. The cassettes add thermal mass to the cycling chamber so 30 cycles for 96 samples takes typically 40 min.
Another method for thermal cycling based on capillary tube resistive thermal cycling has been demonstrated (Friedman and Meldrum 1998). Capillaries externally coated with indium–tin oxide (ITO), a transparent thin resistive film that acts as both a temperature sensor and heater, were shown to perform PCR in 10 min.
Gel Loading
For the results presented here, samples were loaded onto the gels manually, one capillary at a time. As a precursor to high-throughput, automated gel loading, a technique has been developed for high lane density loading onto a horizontal agarose gel directly from the sample capillaries. A simple handheld tool, which mates to the thermal cycling cassettes, was constructed for gel loading. The approach consists of piercing the gel with the pressurized sample capillaries and relieving the pressure shortly before withdrawal. The pressurization prevents the capillary from aspirating the gel buffer and holds the sample at the tip of the capillary so that it may be sucked into the gel during withdrawal. This method has been found to be adequate for PCR-based STS screening and for large and small DNA ladders. In addition to allowing narrower lanes and a higher lane density than is achievable with traditional well-forming techniques, it relaxes the need for well formation and alignment of the sample loader with those wells and provides an easy, efficient means of loading agarose gels (Evensen et al. 1999).
User-Interface and Machine Control
A modular, layered software architecture consisting of a sophisticated graphical user interface, a machine controller, and multiple individual hardware subsystems was designed, built, and implemented on the ACAPELLA-1K system (Arutunian et al. 1998). Each component interacts through a client-server architecture built entirely on top of open Internet standards. The user-interface components are built as Java applets that are downloaded from a server integrated into the computer used to control the instrument (the machine controller). The user-interface client can thereby provide laboratory personnel with a familiar environment for experiment design through a standard World Wide Web browser. Data management and security are seamlessly integrated at the machine-controller layer using QNX, a real-time operating system (QNX Software Systems, Ltd., Kanata, Ontario, Canada). This layer also controls hardware subsystems through a second client-server interface. This architecture has proven flexible and relatively easy to implement and allows users to operate laboratory automation instruments remotely through the Internet. A demonstration of the user interface may be found at the Genomation Laboratory web site (http://rcs.ee.washington.edu/GNl).
PCR Methods
The ACAPELLA-1K system has performed a variety of PCR reactions and restriction enzyme digests. Amplifications reported here were done with human genomic DNA templates from Clontech Laboratories, Inc. (Palo Alto, CA; part no. 6550-1) and the National Institute of General Medical Sciences Cell Repository (Coriell Cell Repositories, Camden NJ; DNA samples NA07057, NA06990B, and NA10848). In addition, PCR was performed on BAC clones from human chromosome 7 provided by the UWGC, and pET21 vector (Novagen Inc.) with and without insert provided by Professor Stayton's laboratory at the UW. The human DNA was probed with various chromosome seven sequence-tagged site (STS) primers, selected because of their use at the UWGC. (GenBank accession nos.G05166, G00083, and G12522) Additionally, a DNA length polymorphism 5′ to the human ATIII gene (Bock and Levitan 1983) caused by the presence of nonhomologous nucleotide sequences was amplified. These variable segments exist in heterozygous and both homozygous forms in the National Institute of General Medical Sciences templates above. The following primers were used to give 143- or 218-bp amplicons: 5′-CTCTCCCTCTCTCCATAAAGAAAAC-3′ and 5′-CCTCACTCTTCTCCTCAGCTTTAT-3′. Finally, the presence or absence of an insert in the pET21 vector was probed with T7 terminator and promoter primers (Novagen, Inc. nos. 69348-3 and 69337-1). All reactions were performed in a RapidCycler capillary thermocycler (Idaho Technology Inc.) at the following parameters: Denature at 94°C with 0-sec hold time, anneal at the appropriate temperature with a 5-sec hold, and extend at 72°C for 15 sec. Reaction products were loaded into standard wells on a 2% agarose gel prepared with 1 μl of ethidium bromide per 100 ml of gel solution.
The 2-μl PCR reactions used on the ACAPELLA-1K system consisted of 2.5 ng of template DNA, 160 μm each dNTP, 0.1 μm each primer, 0.1 unit of Taq DNA polymerase (Promega Corp., Madison, WI), 10× reaction buffer with 20 mm Mg+2 and 2.5 mg/ml BSA (Idaho Technology Inc.), “5×” diluent (10×, 2.5 mg/ml BSA, Idaho Technology Inc.), and an additional 2.25 mg of nonacetylated BSA (20 mg/ml; Roche Bioscience, Palo Alto, CA) to yield a total BSA concentration in the reaction of 3 mg/ml. Reactions prepared for thermal cycling in chambers with high surface-to-volume ratios such as glass capillary tubes and silicon chips generally require 0.5 mg/ml BSA (Wittwer et al. 1990, Taylor et al. 1997); however, the use of the dispenser has required a higher concentration. Recent unpublished work in our group has reduced the required BSA concentration to 0.5 mg/ml by using Amplitaq FS polymerase (Perkin-Elmer Corp., Norwald, CT) (Mohan Saini, unpubl.).
Currently, typical ACAPELLA-1K PCR reactions are carried out using two dispensers. One is filled with 50 μm primer stock and the 2 mm dNTP stock plus water, and the other contains water and all other ingredients besides template DNA. For a PCR reaction protocol on the ACAPELLA-1K system, a fixed DNA volume (550 nl) is first aspirated from the sample tray into a capillary tube. The sample capillary then receives 290 nl from the primer/dNTP dispenser and 1160 nl from the Taq/BSA dispenser for a 2-μl reaction volume.
This distribution of the reagents among the dispensers results in theTaq polymerase being diluted by a factor of 60 from its stock concentration. This is done to reduce the amount of Taq that is lost to the dispenser's dead volume, which is the minimum volume at which the dispenser still delivers a uniform drop size. Because a dead volume of roughly 10 μl per dispenser is lost on the ACAPELLA-1K system, diluting the Taq reduces the number of units that are wasted in a run. This dilution was not found to adversely affect results.
Sequencing Reaction and Purification Methods
Sequencing reactions have been prepared in the system using pGEM DNA and human genomic DNA from the UW Genome Center. The steps for aspirating DNA, adding reagents, mixing, and thermal cycling are the same as those used for PCR. Dye primer sequencing reactions are prepared using Amersham ET-dye primer reagents. Two microliters are prepared per capillary. The reactions are not purified but are pooled (manually) and 2 μl is loaded per lane. Products were sequenced on a PE 373 sequencer in the UW Genome Center. Dye terminator reactions are prepared using PE Big Dye Terminator reagents. After thermal cycling the reactions in glass capillaries in an Idaho Technology air thermal cycler, the products are purified and sequenced on a PE 377 sequencer (8 hr, 2400 V, 52°C, 46 cm length, 4% acrylamide gel with bisacrylamide cross-linker 1:19) in the UWGC.
To purify dye terminator sequencing reactions before sequencing, we developed and demonstrated a proof-of-principle method for purifying products with magnetic beads inside of 5-μl glass capillaries. The method uses 2.8 μm of Dynal streptavidin magnetic beads and primers biotinylated at the 5′ terminal. Suspended magnetic beads and binding buffer are added to a capillary containing 2 μl of sequencing reaction. After the solution is mixed it is held for 5 min to complete the binding of the DNA to the magnetic beads. The solution is separated from the beads by placing the capillaries in a magnet for one min. The supernatant is decanted with a pipette. With the capillary still in the magnetic field, 70% ethanol solution is flushed through to wash the beads. Again, the supernatant is decanted with a pipette. A loading buffer (83% formamide, 8.3 mm EDTA) containing tracking dye is added to the beads in the capillary. The DNA is removed from the magnetic beads by heating at 88°C for 3–4 min. The product is then ready for sequencing. Development of a fully automated version of this purification procedure is currently in progress.
Acknowledgments
We thank the students in the Genomation Laboratory (Ethan Arutunian, Marc Borden, Eric Dixon, Neal Friedman, Wei Hai, Sabine Kunig, Vitaliy Mosesov, Joel Reiter, and Lauren Sjoboen) who have performed experiments on the ACAPELLA system and contributed to various aspects of its development; Dr. Mohan Saini, for performing the sequencing and contamination studies; Dr. Deborah Nickerson, for constructive feedback and her aid on the ATIII PCR primers and templates; and the laboratory of Dr. Patrick Stayton, for access to plasmid samples. We are grateful to Dr. Maynard Olson and the UWGC for many consultations, helpful discussions, and generous collaborations in running experiments. Finally, we gratefully acknowledge the NIH NHGRI for their financial support of this research (grant no. HG01497).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵4 Corresponding author.
-
E-MAIL deedee{at}ee.washington.edu; FAX (206) 543-3842.
-
- Received March 12, 1999.
- Accepted November 19, 1999.
- Cold Spring Harbor Laboratory Press