High-density haplotyping with microarray-based expression and single feature polymorphism markers in Arabidopsis
Supplementary Figure 7, West et al.
Click on the images to open a new window with a larger high-resolution image.
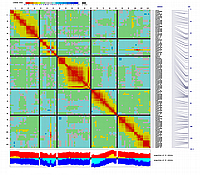
Supplementary Figure 7. GEMs identified with the RIL distribution method. GEMs are arranged in physical order based on the Col-0 genomic sequence.
The five Arabidopsis linkage groups are oriented vertically, laid end-to-end. A) Heat map showing a matrix of recombination scores for GEMs and microsatellite markers assayed on 148 RILs. Pair-wise comparisons
between markers were used to assign recombination scores, which are plotted as a heat map matrix of all markers (top to bottom)
against all markers (left to right). Lowest recombination scores, suggesting marker linkage, are indicated by red boxes,
while blue boxes indicate high recombination scores, suggesting no linkage. Segregation ratios for alleles at each marker
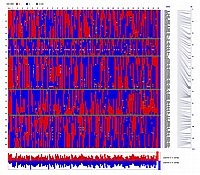
are depicted below the heat map, with red representing Sha alleles and blue representing Bay-0 alleles. B) Haplotypes of 148 RILs plus parental genotypes.
Each column represents a RIL (first 148 columns) or parental genotype (last four columns). Rows correspond to GEMs and microsatellite
markers. Red boxes indicate Sha genotypes, blue boxes indicate Bay-0 genotypes, and grey boxes indicate markers scored as
missing data. The text column (far right) lists the GEM IDs and the 38 microsatellite markers described in Loudet et al.
(2002). The final column depicts the physical distances between the markers. Below the haplotypes, the proportions of Sha
(red bars) and Bay-0 (blue bars) alleles in each RIL are depicted.
This Article
-
Genome Res. June 2006 vol. 16 no. 6 787-795