Whole Genome Comparison of Campylobacter jejuni Human Isolates Using a Low-Cost Microarray Reveals Extensive Genetic Diversity
Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity
Nick Dorrell, 1,4 Joseph A. Mangan, 2,4 Kenneth G. Laing, 2 Jason Hinds, 2 Dennis Linton, 1 Hasan Al-Ghusein, 2 Bart G. Barrell, 3 Julian Parkhill, 3 Neil G. Stoker, 1 Andrey V. Karlyshev, 1 Brendan W. Wren, 1,5 and Philip D. Butcher 2
1 Department of Infectious and Tropical Diseases, London School of Hygiene & Tropical Medicine, London, WC1E 7HT, UK; 2 Department of Medical Microbiology, St George's Hospital Medical School, London, SW17 0RE, UK; 3 The Sanger Centre, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SA, UK.
4 These authors have contributed equally to this work.
5 Corresponding author. Pathogen Molecular Biology and Biochemistry Unit, Department of Infectious and Tropical Diseases, London School of Hygiene & Tropical Medicine, Keppel Street, London, WC1E 7HT, United Kingdom.
Tel: +44 (0)20 7927 2288, Fax: +44 (0)20 7636 8739, E.mail: brendan.wren@lshtm.ac.uk
Abstract
Campylobacter jejuni is the leading cause of bacterial food-borne diarrhoeal disease throughout the world, and yet is still a poorly understood pathogen. This paucity of knowledge on such an important pathogenic bacterium was the driving force behind the recent sequencing of the C. jejuni NCTC 11168 genome. To compare the genome content of C. jejuni strains, we constructed a low-cost whole genome microarray using PCR products amplified with a single pair of vector primers. PCR products representing all 1654 annotated C. jejuni NCTC 11168 genes were amplified from selected clones of an ordered plasmid library of that strain, which was a by-product of the genome sequencing project. Whole genome microarray comparisons of eleven C. jejuni strains of diverse origin identified genes in up to thirty NCTC 11168 loci ranging from 0.7 to 18.7 kb that are either absent or highly divergent in these isolates. Many of these regions are associated with the biosynthesis of surface structures including flagella, lipo-oligosaccharide and the newly identified capsule. Other strain-variable genes of known function include those responsible for iron acquisition, DNA restriction / modification and sialylation. In fact at least 21% of genes in the sequenced strain appear dispensable as they are absent or highly divergent in one or more of the isolates tested, thus defining 1300 C. jejuni core genes. Such core genes contribute mainly to metabolic, biosynthetic, cellular and regulatory processes, but many virulence determinants are also conserved. Comparison of the capsule biosynthesis locus revealed conservation of all the genes in this region in strains with the same Penner serotype as strain NCTC 11168. By contrast, between five and seventeen NCTC 11168 genes in this region are either absent or highly divergent in strains of a different serotype to the sequenced strain, providing further evidence that the capsule accounts for Penner serotype specificity. These studies reveal extensive genetic diversity among C. jejuni strains and pave the way towards identifying correlates of pathogenicity and developing improved epidemiological tools for this problematic pathogen.
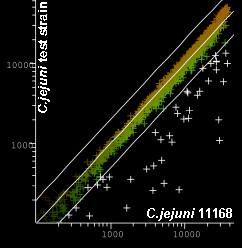
Typical Array Image and Scatter Plot
|
Red / Green false colour image of microarray. Green: C.jejuni 11168 , Red: C.jejuni test strain
|
Scatter plot of C.jejuni 11168 vs. C.jejuni test strain. Genes selected as absent or highly divergent shown in white |
|
 |
 |
Supplementary Data
Individual array data for each of the eleven strains analysed by three independent hybridisations, the lists of absent or highly divergent genes and the data set consisting of background subtracted normalised R/G signal ratios.
N.B. With regard to GeneSpringTM 3.2.12 software analysis of the data, any gene whose R/G signal ratio is below 0.5 but does not appear in the gene deletion list, was excluded if the control signal in the Cy3 channel was less than 100 units (see METHODS; Data acquisition and analysis);
| 81116 | Human isolate (Chelmsford 1981), extensively studied laboratory strain |
| 81-176 | Human isolate (Minnesota 1981), extensively studied laboratory strain |
| X | Human isolate, gastroenteritis (London 1997) |
| G1 | Human isolate, Guillain-Barre syndrome |
| P1 | Penner serotype O:1 reference strain |
| P2 | Penner serotype O:2 reference strain |
| P3 | Penner serotype O:3 reference strain |
| P4 | Penner serotype O:4 reference strain |
| PHLS01 | Human isolate, serotype O:2, phage type 35 (PHLS, UK) |
| PHLS02 | Human isolate, serotype O:2, phage type 1 (PHLS, UK) |
| PHLS03 | Human isolate, serotype O:2, phage type 44 (PHLS, UK) |
Additional Information
- BlastN Scores for single clones representing each gene on the array against annotated C. jejuni NCTC11168 gene sequences
- Composition of single clones representing each gene on the array
For Further Information on annotated C. jejuni NCTC 11168 genes
This Article
-
Genome Res. October 1, 2001 vol. 11 no. 10 1706-1715




