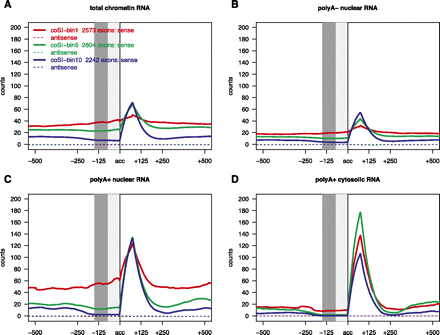
RNA-seq profile plots using RNA-seq reads mapping to the genome only, aligned at the acceptor. At each aligned position, the average number of overlapping RNA-seq reads (mapping to the genome) for all exons in each bin (according to coSI values in the total chromatin-associated RNA fraction in all four subfigures) is plotted for sense (solid lines) and antisense strand (dashed lines). Here, only exons that are at least 150 bp away from any other exon are used. RNA-seq profiles for the total chromatin-associated RNA fraction (A), the polyA− nuclear fraction (B), the polyA+ nuclear fraction (C), and the polyA+ cytosolic fraction (D). (Dark gray area) Positions that are guaranteed to be covered only by reads that were not used for the coSI value calculation; (both gray areas) positions that are guaranteed to be intronic. Note that these profiles are not normalized for gene expression. We added profiles normalized for cytosolic polyA+ gene expression in Supplemental Figure S6.











