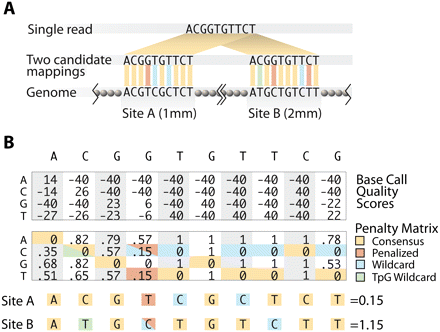
Mapping bisulfite treated reads. (A) Reads were mapped to the reference genome by minimizing the number of potential mismatches. Any T in a read incurred no penalty for aligning with a C in the genome, and any C in a read was penalized for aligning with a T in the genome. (B) Quality scores were converted to mismatch penalties by assigning a penalty of 0 to the consensus base, and penalizing non-consensus bases proportionately to the difference between their quality score and the consensus base score. A difference of 80 (representing the maximum possible range at a single position) was equated with a penalty of 1.











