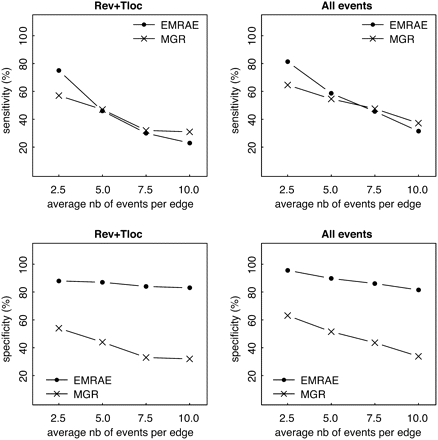
Sensitivity and specificity of EMRAE and MGR in predicting rearrangement events based on simulated data sets using two rearrangement models: Rev+Tloc (reversals and translocations) and All events (reversals, transpositions, translocations, and fusions/fissions). The x-axis corresponds to the evolutionary rate as defined by the average number of events simulated on each edge of the tree. nb, number.











