Robust 16S rRNA classification based on a compressed LCA index
Abstract
Taxonomic sequence classification is a computational problem central to the study of metagenomics and evolution. Advances
in compressed indexing with the r-index enable full-text pattern matching against large sequence collections. But the data structures that link pattern sequences
to their clades of origin still do not scale well to large collections. Previous work proposed the document array profiles,
which use 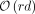 words of space, where r is the number of maximal equal-letter runs in the Burrows–Wheeler transform, and d is the number of distinct genomes. The linear dependence on d is limiting, because real taxonomies can easily contain 10,000s of leaves or more. We propose a method called cliff compression
that reduces this size by a large factor, >250× when indexing the SILVA 16S rRNA gene database. This method uses
words of space, where r is the number of maximal equal-letter runs in the Burrows–Wheeler transform, and d is the number of distinct genomes. The linear dependence on d is limiting, because real taxonomies can easily contain 10,000s of leaves or more. We propose a method called cliff compression
that reduces this size by a large factor, >250× when indexing the SILVA 16S rRNA gene database. This method uses 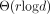 words of space in expectation under a random model we propose here. We implemented these ideas in an open-source tool called
Cliffy that performs efficient taxonomic classification of sequencing reads with respect to a compressed taxonomic index.
When applied to simulated 16S rRNA reads, Cliffy's read-level accuracy is higher than Kraken2's by 11%–18%. Clade abundances
are also more accurately predicted by Cliffy compared with Kraken2 and Bracken. Overall, Cliffy is a fast and space-economical
extension to compressed full-text indexes, enabling them to perform fast and accurate taxonomic classification queries. Cliffy's
accuracy underscores the advantages of full-text indexes, which offer a more precise solution compared with k-mer indexes designed for a specific k value.
words of space in expectation under a random model we propose here. We implemented these ideas in an open-source tool called
Cliffy that performs efficient taxonomic classification of sequencing reads with respect to a compressed taxonomic index.
When applied to simulated 16S rRNA reads, Cliffy's read-level accuracy is higher than Kraken2's by 11%–18%. Clade abundances
are also more accurately predicted by Cliffy compared with Kraken2 and Bracken. Overall, Cliffy is a fast and space-economical
extension to compressed full-text indexes, enabling them to perform fast and accurate taxonomic classification queries. Cliffy's
accuracy underscores the advantages of full-text indexes, which offer a more precise solution compared with k-mer indexes designed for a specific k value.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279846.124.
- Received July 24, 2024.
- Accepted August 7, 2025.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see https://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.











