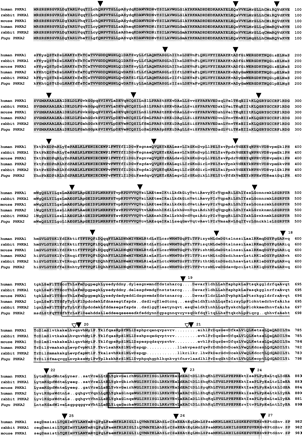

Alignment of Fugu, rabbit, and human PHKA2 and mouse, rabbit, and human PHKA1 proteins. The alignment was performed with the GCG programs PILEUP and PRETTY. Residues identical at each position are shown in uppercase letters and are shaded. (▾) Intron/exon boundaries of Fugu PHKA2. Fugu exons 18–29 are numbered. (▿) Known positions of splice sites in the humanPHKA2 gene. The PHKA muscle and liver isoforms have two subunit-specific domains (boxed) with comparatively low amino acid sequence similarities of <50%. These domains encompass amino acids 612–788 and amino acids 1027–1055 in Fugu with a negatively charged cluster spanning amino acids 620–645. The hypothetical 5′ and 3′ calmodulin binding sites of all Phk α subunits are highly conserved (boxed in bold) and encompass amino acids 828–855 and amino acids 1056–1087 in Fugu. Three autophosphorylation sites, serine 987, serine 1001, and serine 1022, labeled by an P underneath the alignment and their vicinity are conserved.











