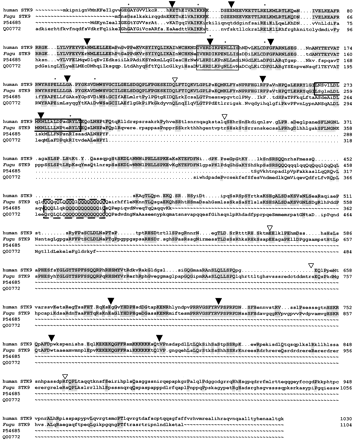
Multiple alignment of the human, Fugu, Dictyostelium(P54685), and yeast (Q00772) STK9 proteins. The alignment was performed with the GCG programs PILEUP and LINEUP. Amino acids identical in at least two proteins are shown in uppercase letters, shading indicates residues identical in Fugu and human and in all proteins, respectively. (▾) Intron/exon boundaries identical in Fuguand human STK9; (▿) splice sites only present inFugu. The protein kinase ATP-binding region is presented by the first box. The second box indicates the predicted serine-threonine protein kinase active site. The predicted leucine zipper is boxed in bold and the polyglutamine stretch in broken lines. (Asterisks) Amino acid residues invariant in 60 members of the eukaryotic protein kinase superfamily (Hanks and Hunter 1995).











